Академический Документы
Профессиональный Документы
Культура Документы
Project 3
Загружено:
crickel101Исходное описание:
Авторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Project 3
Загружено:
crickel101Авторское право:
Доступные форматы
Chelsea Rickel November 27, 2012
Linear Equations and Least Squares
In our third project, we implemented Gaussian elimination with two methods for solving linear systems with nonsingular matrices, and also compared the total error difference between linear and exponential models when using the least squares method. Through this project, we will see which methods are preferred and why. Determining the correct method will ensure we receive suitable results. For the first phase of our project, we implemented nave Gaussian elimination and Gaussian elimination with partial pivoting in Matlab. The two differ because nave Gaussian elimination makes assumptions about the matrix A, which is passed in as an input variable. The code I implemented was based on the Matlab code we covered in lecture when discussing the methods of Gaussian elimination. To control which method was used, I added if statements which check the method input variable for a value equal to zero, or if not equal to zero. If the value is equal to zero, the nave method is used. The nave method assumes the matrix A is a nonzero matrix, meaning there is no possible scenario where we will be dividing by zero. There is no row swapping with the nave method because the main reason to swap rows is when there is a conveniently placed zero that would help create a triangular matrix easier. If the value of the method input is not zero, the partial pivoting method will be used instead. Partial pivoting takes advantage of its ability to swap rows since this method does not assume A to be a non-zero matrix. Additionally, I added a check to see if the matrices were compatible, and if not the
x matrix was transposed. To test both methods, I used examples from the textbook on page 247 and page 265, and received the correct results. After confirming that I was receiving the correct output for both textbook examples, I tested the Hilbert matrix file provided in the project description. The Hilbert matrix grows particularly quickly, and can become very difficult to work with. The matrix size, defined as (n, n), is dependent on the variable i. As i increases, n increases by 50 times that. For instance, when i = 1 the matrix size is (50, 50), and when i = 2 the matrix size is (500, 500). It is clear that the matrix can become rapidly large with each iteration. The main focus of phase one of our project was to compare the residual error of the matrix to the solution error using the two methods mentioned previously. The residual error is based on Ax = b, and the solution error is based off x = xtrue. This can be seen in the Matlab code. In the case of the Hilbert matrix, the residual error was less than the solution error. This is because in the case of the Hilbert matrix test file we are solving for Ax = b, not x = xtrue. Interestingly enough, we can also see that the error in the residual grows by a factor of ten through the iterations and the solution error grows by a factor of one thousand. For the second phase of our project, we used the least squares method to determine the approximate solution to a set of functions. The first function we found an approximate solution for was a linear function. We were provided with the Matlab code necessary to determine the model RMS error. The second function we found an approximate solution for was an exponential function. The Matlab code for determining the error for this function was similar to the linear function code, but
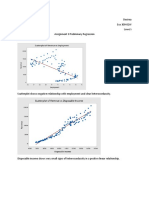
had significant differences. First, I changed the function y in the exp_eval file to an exponential function that was defined in the project description. Second, I adjusted the error if statement in exp_eval to be less than three because the function y has three c values. In the exp_solve we create the least squares matrix, and when the function is adjusted the creation of the matrix must be adjusted as well. Matrix A will still have as many rows equal to the length of x, but the number of columns will increase to three. Also, each column of A needs to be adjusted based on the new equation. Now that the exponential least squares code is corrected, we can compare the model RMS error, which is the total error of the model against the data points. Between the two functions, the error for the linear equation is slightly less than the error for the exponential equation. This means the linear model is better matched with the data provided in the file. The graphs from the linear equation and exponential equation are below.
For the last phase of our project, we are given a climate data file with a significant amount of noise due to yearly variation. The noise prevents us from simply connecting the dots. We must instead define a function that will represent a general idea of the path the data points draw. We must use the least squares method to create a model of the data, in hopes of being able to predict what climate we can expect in the future. The first step in phase three of our project is to use the exponential model from the previous phase to determine a general model for the climate data. After seeing the model RMS error associated with the generated model, it is our task to attempt to improve that error by reducing the value to be as close to zero as we can. My approach to this task was to choose a function with degree five. I tested polynomials with a degree higher and lower than five, sine and cosine functions, and logarithmic functions, but a polynomial with degree five provided the lowest model RMS error. This error is also slightly lower than the exponential functions error from the earlier modeling of the climate data, which means my equation matches better with the data points than the exponential equation. When observing the graph formed by the data points and model, the function of my choice seems to represent the data points fairly well. I would rather have a lower error than this function provides, but after comparing errors from multiple functions a polynomial of degree five provides the lowest error of the bunch. The future climates from this function, on the other hand, are possibly represented worse than the exponential function. There is no sure way to determine the climate of fifty years into the future, but with the least squares method, and a model that
flows through current climate data points, we can attempt to predict future information. Unfortunately, though my function lowers the model RMS error of the current climate, the error for future climates seem to be increased. The chances of such a drastic increase in temperature are very unlikely. A better representation of possible future climates can be formed with a more complicated function, which includes a cosine, logarithmic, and square root function. In this case, future climates stay around 58 degrees, with only a mild increase of temperature over time. The down side to using this equation is that the model RMS error is even larger than the exponential functions error. For this reason, I favor the polynomial with degree five to represent the climate data file. The two options of functions are graphed below.
After observing the result of each method demonstrated in this project, we can confirm that some methods are more useful than others. First, partial pivoting benefits us more than the nave method by not assuming anything about the given input. Gaussian elimination with partial pivoting allows the user to benefit from abilities, such as swapping rows, that nave Gaussian elimination will not take into
account. Secondly, modeling data can be tricky, but because we aim to lower the model RMS error to be as close to zero as possible we are able to target a function that will give us just that. Lastly, it is difficult to predict what the future may bring us, but we can use the least squares method to attempt to determine a possible outcome of the future, as we did with climates. With these considerations, I feel confident of my ability to choose a worthy method when dealing with linear equations and least squares.
gauss_elm.m function [x] = gauss_elm(A, b, method) if(method == 0) % naive gauss_elm P252 % tested with P247 matrices and worked! xmult = 0; sum = 0; n = length(A); % small pivot elements lead to large for k=1:n-1 % multipliers and to worse roundoff for i=k+1:n % errors xmult = A(i,k)/A(k,k); A(i,k) = xmult; for j=k+1:n % triply nested for-loop containing replacement operator A(i,j) = A(i,j) -(xmult*A(k,j)); end b(i) = b(i) - (xmult*b(k)); end end x(n) = b(n) / A(n,n); for i=n-1:-1:1 sum = b(i); for j=i+1:n sum = sum - (A(i,j)*x(j)); end x(i) = sum / A(i,i); end x = x'; end if(method ~= 0) % gauss_elm with partial pivoting P267 % tested with P247 matrices and worked! xmult = 0; sum = 0; n = length(A); for k=1:n-1 p = k; for i=k+1:n if(abs(A(i,k)) > abs(A(p,k))) p = i; end end temp = b(k); b(k) = b(p); b(p) = temp; temp = A(k,k:n);
A(k,k:n) = A(p,k:n); A(p,k:n) = temp; if (A(k,k) == 0) fprintf('Error: A(k,k) = 0') end for i=k+1:n % errors xmult = A(i,k)/A(k,k); A(i,k) = xmult; for j=k+1:n % triply nested for-loop containing replacement operator A(i,j) = A(i,j) -(xmult*A(k,j)); end b(i) = b(i) - (xmult*b(k)); end end x(n) = b(n) / A(n,n); for i=n-1:-1:1 sum = b(i); for j=i+1:n sum = sum - (A(i,j)*x(j)); end x(i) = sum / A(i,i); end x = x'; % deallocate array s(i) end end
exp_eval.m function y = exp_eval(c,x) % Usage: y = exp_eval(c,x) % % Chelsea Rickel % Math 3315 / CSE 3365 % Fall 2012 % check that there are enough coefficients if (length(c) < 3) error('exp_eval error: input vector c is too short (minimum length 3)') end % evaluate the linear model % g(x) = ec0 x2 +c1 x+c2 y = exp(c(1)*x.^2 + c(2)*x + c(3)); % end the function end
exp_solve.m function c = exp_solve(x,f) % Usage: c = exp_solve(x,f) % Chelsea Rickel % Math 3315 / CSE 3365 % Fall 2012 % check that the input data sets are the same size if (length(x) ~= length(f)) error('exp_solve error: input vectors are not the same length') end % get the number of data points n = length(x); % ensure that x and f are column vectors x = reshape(x,n,1); f = reshape(f,n,1); % create the least-squares matrix A = zeros(n,3); % our equation has max degree of 3, so A needs 3 columns A(:,1) = x.^2; A(:,2) = x; A(:,3) = ones(n,1); % create the right-hand side vector b = log(f); % because our equation being evaluated in an exponent, we need log of f % solve the least-squares system for the coefficients c c = (A'*A)\(A'*b); % end the function end
exp_test.m % exp_test.m: % Chelsea Rickel % Math 3315 / CSE 3365 % Fall 2012 % start with a clean slate clear % input the data set data = load('data_exp.txt'); % extract the x and f arrays from this data set x = data(:,1); f = data(:,2); % set the number of data nodes n = length(x); % call lin_solve to generate the modeling coefficients c = exp_solve(x,f); % evaluate model at all input values g = exp_eval(c,x); % compute RMS norm of the modeling error totalerror = sqrt(sum((f-g).^2)/n); fprintf(' Model RMS error = %g\n',totalerror); % set plotting interval xmin = min(x); xmax = max(x); xvals = linspace(xmin, xmax, 1000); % evaluate model at all points in plotting interval gvals = exp_eval(c,xvals); % overlay plots of the data set and the model figure() plot(x,f,'c*',xvals,gvals,'b-') legend('data','model') xlabel('x') ylabel('f(x),g(x)') title('Data and Exponential Model') % end of script
exp_climate.m % exp_climate.m: % % Chelsea Rickel % Math 3315 / CSE 3365 % Fall 2012 % start with a clean slate clear % input the data set data = load('data_climate.txt'); % extract the x and f arrays from this data set % there are two columns, each column is set to a variable x = data(:,1); f = data(:,2); % set the number of data nodes n = length(x); % call lin_solve to generate the modeling coefficients c = exp_solve(x,f); % evaluate model at all input values g = exp_eval(c,x); % compute RMS norm of the modeling error totalerror = sqrt(sum((f-g).^2)/n); fprintf(' Model RMS error = %g\n',totalerror); % set plotting interval xmin = min(x); xmax = max(x); xvals = linspace(xmin, xmax, 1000); % evaluate model at all points in plotting interval gvals = exp_eval(c,xvals); % overlay plots of the data set and the model figure() plot(x,f,'c*',xvals,gvals,'b-') hl = legend('data', 'model', 'location', 'NorthWest'); %legend('data','model')
xlabel('x') ylabel('f(x),g(x)') title('Data and Exponential Model') hold on T2015 = exp_eval(c, 2015) T2025 = exp_eval(c, 2025) T2050 = exp_eval(c, 2050) plot(2015, T2015, 'ro') plot(2025, T2025, 'ro') plot(2050, T2050, 'ro') % NEED TO UPDATE LEGEND hold off % end of script
my_eval.m function y = my_eval(c,x) % Usage: y = my_eval(c,x) % % Chelsea Rickel % Math 3315 / CSE 3365 % Fall 2012 % check that there are enough coefficients if (length(c) < 6) error('my_eval error: input vector c is too short (minimum length 6)') end % evaluate the linear model %y = exp(c(1)*(sin(x)).^2 + c(2)*log(x) + c(3)*sqrt(x)); y = c(1)*x.^5 + c(2)*x.^4 + c(3)*x.^3 + c(4)*x.^2 + c(5)*x + c(6); %y = c(1)*x.^6 + c(2)*x.^5 + c(3)*x.^4 + c(4)*x.^3 + c(5)*x.^2 + c(6)*x + c(7); % end the function end
my_solve.m function y = my_eval(c,x) % Usage: y = my_eval(c,x) % % Chelsea Rickel % Math 3315 / CSE 3365 % Fall 2012 % check that there are enough coefficients if (length(c) < 6) error('my_eval error: input vector c is too short (minimum length 6)') end % evaluate the linear model %y = exp(c(1)*(sin(x)).^2 + c(2)*log(x) + c(3)*sqrt(x)); y = c(1)*x.^5 + c(2)*x.^4 + c(3)*x.^3 + c(4)*x.^2 + c(5)*x + c(6); %y = c(1)*x.^6 + c(2)*x.^5 + c(3)*x.^4 + c(4)*x.^3 + c(5)*x.^2 + c(6)*x + c(7); % end the function end
my_climate.m % exp_climate.m: % % Chelsea Rickel % Math 3315 / CSE 3365 % Fall 2012 % start with a clean slate clear % input the data set data = load('data_climate.txt'); % extract the x and f arrays from this data set x = data(:,1); f = data(:,2); % set the number of data nodes n = length(x); % call lin_solve to generate the modeling coefficients c = my_solve(x,f); % evaluate model at all input values g = my_eval(c,x); % compute RMS norm of the modeling error totalerror = sqrt(sum((f-g).^2)/n); fprintf(' Model RMS error = %g\n',totalerror); % set plotting interval xmin = min(x); xmax = max(x); xvals = linspace(xmin, xmax, 1000); % evaluate model at all points in plotting interval gvals = my_eval(c,xvals); % overlay plots of the data set and the model figure() plot(x,f,'c*',xvals,gvals,'b-') hl = legend('data', 'model', 'location', 'NorthWest'); %legend('data','model') xlabel('x') ylabel('f(x),g(x)')
title('Data and My Model') hold on T2020 = my_eval(c, 2020) T2030 = my_eval(c, 2030) T2040 = my_eval(c, 2040) T2050 = my_eval(c, 2050) plot(2020, T2020, 'ro') plot(2030, T2030, 'ro') plot(2040, T2040, 'ro') plot(2050, T2050, 'ro') % NEED TO UPDATE LEGEND hold off % end of script
Вам также может понравиться
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceОт EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceРейтинг: 4 из 5 звезд4/5 (895)
- Never Split the Difference: Negotiating As If Your Life Depended On ItОт EverandNever Split the Difference: Negotiating As If Your Life Depended On ItРейтинг: 4.5 из 5 звезд4.5/5 (838)
- The Yellow House: A Memoir (2019 National Book Award Winner)От EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Рейтинг: 4 из 5 звезд4/5 (98)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeОт EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeРейтинг: 4 из 5 звезд4/5 (5794)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaОт EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaРейтинг: 4.5 из 5 звезд4.5/5 (266)
- The Little Book of Hygge: Danish Secrets to Happy LivingОт EverandThe Little Book of Hygge: Danish Secrets to Happy LivingРейтинг: 3.5 из 5 звезд3.5/5 (400)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureОт EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureРейтинг: 4.5 из 5 звезд4.5/5 (474)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryОт EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryРейтинг: 3.5 из 5 звезд3.5/5 (231)
- The Emperor of All Maladies: A Biography of CancerОт EverandThe Emperor of All Maladies: A Biography of CancerРейтинг: 4.5 из 5 звезд4.5/5 (271)
- The Unwinding: An Inner History of the New AmericaОт EverandThe Unwinding: An Inner History of the New AmericaРейтинг: 4 из 5 звезд4/5 (45)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersОт EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersРейтинг: 4.5 из 5 звезд4.5/5 (345)
- Team of Rivals: The Political Genius of Abraham LincolnОт EverandTeam of Rivals: The Political Genius of Abraham LincolnРейтинг: 4.5 из 5 звезд4.5/5 (234)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreОт EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreРейтинг: 4 из 5 звезд4/5 (1090)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyОт EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyРейтинг: 3.5 из 5 звезд3.5/5 (2259)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)От EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Рейтинг: 4.5 из 5 звезд4.5/5 (121)
- ASTM G 16 - 95 r99 - Rze2ltk1ujk5rteДокумент14 страницASTM G 16 - 95 r99 - Rze2ltk1ujk5rteCordova RaphaelОценок пока нет
- Experimentation - An Introduction To Measurement Theory and Experiment Design D C BairdДокумент210 страницExperimentation - An Introduction To Measurement Theory and Experiment Design D C BairdCarlos VillalobosОценок пока нет
- Lecture 7 - CH 3 Forecasting - 1sppДокумент58 страницLecture 7 - CH 3 Forecasting - 1sppsatyavaniОценок пока нет
- Chapter 1Документ22 страницыChapter 1Adilhunjra HunjraОценок пока нет
- Nama: Irda Puspasari NPM: 12-087: RegressionДокумент2 страницыNama: Irda Puspasari NPM: 12-087: RegressionAnonymous qBsWolDnoОценок пока нет
- Notes ch3 Sampling DistributionsДокумент20 страницNotes ch3 Sampling DistributionsErkin DОценок пока нет
- 1291TAManual F16Документ134 страницы1291TAManual F16fatjonОценок пока нет
- Business Statistics Midterm Exam: Fall 2019: BUS41000Документ16 страницBusiness Statistics Midterm Exam: Fall 2019: BUS41000vinayak mishraОценок пока нет
- ML ExamДокумент5 страницML ExamAravind Kumar ReddyОценок пока нет
- Best Paper To Learn About ArdlДокумент27 страницBest Paper To Learn About Ardlsaeed meo100% (7)
- Leadership Styles and Employee Performance: Empirical Evidence From Selected Banks in Edo StateДокумент15 страницLeadership Styles and Employee Performance: Empirical Evidence From Selected Banks in Edo StatemeseretОценок пока нет
- Saurav ExamДокумент9 страницSaurav ExamShrijan BasnetОценок пока нет
- Accounting 202 Chapter 6 NotesДокумент13 страницAccounting 202 Chapter 6 NotesnitinОценок пока нет
- Desirea Eco 309-02W Lowe's Assignment 3 Preliminary RegressionДокумент5 страницDesirea Eco 309-02W Lowe's Assignment 3 Preliminary RegressionMEGATRONОценок пока нет
- BS 812-Part 101-84Документ8 страницBS 812-Part 101-84anish_am2005100% (2)
- Imperatives For Modelling and Forecasting Petroleum Products Demand For NigeriaДокумент14 страницImperatives For Modelling and Forecasting Petroleum Products Demand For NigeriaLintang CandraОценок пока нет
- CH 9Документ35 страницCH 9Yoga PratamaОценок пока нет
- Forecasting: Types of Forecasting ModelsДокумент8 страницForecasting: Types of Forecasting Modelslovely abinalОценок пока нет
- A Model of True Spreads On Limit Order Markets: James Mcculloch April 20, 2011Документ28 страницA Model of True Spreads On Limit Order Markets: James Mcculloch April 20, 2011danilam2Оценок пока нет
- Unit 9Документ16 страницUnit 9swingbikeОценок пока нет
- Modeling Risk and Realities Week 4 Session 3Документ25 страницModeling Risk and Realities Week 4 Session 3hiteshОценок пока нет
- Replyto (Cpu Calculations For True Position With MMC Modifier)Документ4 страницыReplyto (Cpu Calculations For True Position With MMC Modifier)Krunal PandyaОценок пока нет
- Lesson 9.1.2 - NoteДокумент9 страницLesson 9.1.2 - NoteFatemeh YarahmadiОценок пока нет
- MSBVARДокумент92 страницыMSBVAREstefania Uribe GaviriaОценок пока нет
- Machine Learning Project Basic - Linear Regression - Kaggle PDFДокумент10 страницMachine Learning Project Basic - Linear Regression - Kaggle PDFKarthikRaoОценок пока нет
- Aircraft Gas Turbine Engine Health Monitoring SystДокумент13 страницAircraft Gas Turbine Engine Health Monitoring SystWilliam Jaldin CorralesОценок пока нет
- Marketing Analytics - Group Assignment 1Документ3 страницыMarketing Analytics - Group Assignment 1SIDDHARTH SOMESHОценок пока нет
- Quantitative Analysis For Management Ch04Документ71 страницаQuantitative Analysis For Management Ch04Qonita Nazhifa100% (1)
- Bias Variance Ridge RegressionДокумент4 страницыBias Variance Ridge RegressionSudheer RedusОценок пока нет
- Unit 4 Research DesignДокумент20 страницUnit 4 Research Designgosaye desalegnОценок пока нет