Академический Документы
Профессиональный Документы
Культура Документы
Hekix Sheet
Загружено:
Parijat BanerjeeАвторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Hekix Sheet
Загружено:
Parijat BanerjeeАвторское право:
Доступные форматы
Proc. Nati. Acad. Sci. USA Vol. 74, No. 10, pp.
4130-4134, October 1977 Chemistry
Structure of proteins: Packing of a-helices and pleated sheets
(close packing/preferred conformations/protein secondary and tertiary structure)
CYRUS CHOTHIA*, MICHAEL LEVITTt,
*
AND
DOUGLAS RICHARDSON*
William Ramsay, Ralph Forster, and Christopher Ingold Laboratories, University College London, Gower Street, London, W.C.1., England; and t Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge, England
Communicated by Linus Pauling, July 22, 1977
ABSTRACT Simple models are presented that describe the rules for almost all the packing that occurs between and among a-helices and pleated sheets. These packing rules, together with the primary and secondary structures, are the major determinants of the three-dimensional structure of proteins.
Twenty-six years ago, Pauling and his colleagues (1, 2) presented the a-helix and the parallel and antiparallel pleated sheets as models for the local folding (secondary structure) of the polypeptide chain in proteins. Since then, various physical techniques (principally x-ray crystal structure analysis) have clearly shown that these secondary structures are almost universally present in protein molecules. We present here models that describe the rules that govern how a-helices and pleated sheets pack together to form the three-dimensional (tertiary) structure of proteins. The rules were developed empirically: a priori models were checked and refined by a detailed analysis of the residue-to-residue contacts that occur between and among the a-helices and pleated sheets in 17 proteins. To do this analysis we made extensive use of a computer graphics system for proteins developed by P. J. Pauling and his colleagues (to be published) and of numerical calculations. The models describe almost all of the secondary structure packings we have so far observed. In a later publication they will be used to present a detailed analysis of known structures. The determining principles Two principles have a dominating influence on the way in which secondary structures associate. 1. Residues that become buried in the interior of a protein close-pack: they occupy a volume similar to that which they occupy in crystals of their amino acid (3, 4). 2. Associated secondary structures retain a conformation close to the minimum free energy conformation of the isolated secondary structures. This second principle is illustrated by the observation that almost all the protein main-chain torsion angles, sp and ,6, lie in regions of torsion angle space that are free from steric strain that is, in the normally allowed regions of the Ramachandran map (5). Le Master and Richards, as reported by Richards (6), found that this is also true for the side-chain angle xi. In a detailed analysis of the conformational potentials of the residue side chains in bovine pancreatic trypsin inhibitor, Gelin and Karplus (7) showed that their conformations were close to those of the free amino acid. The static close-packed image of proteins that is implicit in these two principles is, of course, only true of the time-averaged structure. Individual molecules are subject to large transient thermodynamic (and therefore conformational) fluctuations
The two principles imply that the secondary structures found in protein molecules interact in a manner that gives the maximum van der Waals energy and induces no appreciable steric strain. The rules described below for secondary structure associations arise from these two principles and from the intrinsic geometrical properties of a-helices and pleated sheets.
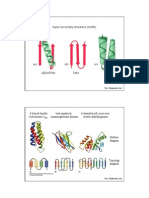
Helix-helix packing To illustrate our model for helix-helix packing, we use a graphical construction invented by Crick (9). The residue side chains in a helix with 3.6 residues per turn and radius 5 A are assumed to have the same size and shape. Two helices are slit down one side in a direction parallel to their axis. Each is opened up and laid flat, one helix face down and one face up. Placing the first lattice over the second is equivalent to bringing the outside of the two helices into contact (Fig. 1). The model for helix-helix interactions is shown in Fig. 1. The surface of a helix can be described in terms of rows of adjacent side chains. A residue in a helix, i, has two neighbors above it, i + 3 and i + 4, and two neighbors below it, i -3 and i -4. One row is formed by the residues i, i i 3, i i 6, . . i i 3n and another by i, i 4, i i 8,. .. i + 4n. The alignment of the side chains so they point to their +3 or 4 neighbors means that the i 3n or i 4n residue rows form a ridge. The similar alignment of adjacent residues means in turn that the helix surface consists of one of two series of parallel ridges separated by shallow grooves: one series being formed by the rows i 3n, (i i 1) i 3n, . . and the other by the rowsi + 4n, (i + 1) 4n, .... Our model for helix-helix packing requires that the surface ridges in the first helix pack into the grooves between the ridges in the second helix and vice versa (Fig. 1). As is apparent from Fig. 1, this model gives three classes of interaction that differ in the residues they bring into contact and in the angle between the helix axes (Q)t (Table 1). Real helix-helix packings will deviate from this simple model for two reasons. First, the angle between the helix axis and the rows of residues will depend upon the exact twist and radius of the helix. Thus, if the two ideal helices of radius 5 A each have 3.4 residues per turn, the Q values will be -105, -81, and -3, and if they each have 3.8 residues per turn the Q values will be -66, -32, and +40. Second, the side chains do not all have the same size and shape but vary between 0 and 10 atoms. From the atomic coordinates we have calculated the contacts that occur between the helices and in the a subunit of hemoWe use Q to describe the relative orientation of two pieces of secondary structure in contact. Q is defined as the angle between the strands of the pleated sheet and/or helix axes when projected onto their plane of contact. We ignore the direction of individual a-helices and strands so 0 is defined between -90 and +90 rather than -180 and +180. The angle is negative (0 -- -900) if the near helix or strand is rotated in a clockwise direction relative to the far helix or strand. If this rotation is anticlockwise, the angle is positive (0 +900).
(8).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 U. S. C. 1734 solely to indicate this fact.
4130
Chemistry: Chothia et al.
Proc. Natl. Acad. Sci. USA 74 (1977)
CLASS I
127
10
4131
128
125
12
Hb
FACE
UP FAC E DOWN
CLASS II
b
CLASS I
i 3n
a
CLASS II it4n j4n
C
CLASS m i4n jI3n
10 76
148
77 144 %s4 145 l42
141
-9Oo
137
jt3n
8x
TLS
CLASS III
OO00
115
0000
0
141
142
027
104
kI3!91.o 39
SUB
132
180
nu -82
As-60
fQ
FIG. 2. The observed helix-helix packing in the a-subunit of hemoglobin (Hb), thermolysin (TLS), lysozyme, staphylococcal nuclease, calcium-binding protein, and subtilisin (SUB) (see text). We show Q for the 25 observed helix-helix packings and one example, for each class, of the residue-residue contacts: 0, side chains on the far helix; X, side chains of the near helix. Lines joining the symbols indicate the residue-residue contacts (see also Fig. Id).
d
FIG. 1. The model for helix-helix packing: The residues in a helix (a) are represented by large circles. This helix is opened up to give two flattened projections (see text) shown face-up in b and face-down in c. The residue rows i A 3n, i A 4n, j 3n and j 4n are labeled. (d) The three classes of "ridges into grooves" packing. The Q values are for two ideal helices with 3.6 residues per turn and radius 5 A. Lighter lines indicate the j 3n and j + 4n rows of residues.
Inspection by computer graphics of the six class III interactions shows that three of them have a systematic deviation from
globin, thermolysin, lysozyme, calcium-binding protein, subtilisin, and staphylococcal nuclease. We found 26 cases of helix-helix packing in which three or more residues from one helix are in contact with three or more residues from the other. We have also calculated, for each of the 26 pairs, the angle between the helix axes. Twenty-five of these can be related to our moder for helix-helix packing (in the 26th case, the helices are joined covalently by a disulfide bridge). Inspection of the pattern of residue contacts shows that 3 of these interactions are of a type we have defined as class I, 16 are class II, and 6 are class III. For class I, the observed values of Q are -80, -850, and -950. In class II the angles vary between -20 and -70, although 11 are in the range -50 + 100 (Fig. 2).
Table 1. Classes of interaction Residues forming rows Q* First helix Second helix I i i 3n ]i3n -820 II i 4n j i 4n -600 III i +4n +190 i3n * Angle between helix axes, for two helices of 3.6 residues per turn and radius 5 A. Class
the simple model with Q close to 00 and each helix tilted about 100 away from the plane of contact. Details of this will be published later. The other three members of this class have Q of +50, +150, and +350 (Fig. 2). The 25 helix-helix packings involve 50 different helix surfaces. Thirty-eight of these have the ridges formed by the i i 4n residue rows. This is due to the side chain, i, preferring to point toward the i - 4 neighbor. The model for helix-helix packing presented here differs from that previously described by Crick (9). In his model, residue i from one helix fitted in between residues j, j + 3, j + 4, and j + 7 in the second helix and vice versa. This gives two packing patterns in which the helix axes are inclined at +200 or -70. Although a few of the helix interactions observed here do occur at angles close to these values, the pattern of residueresidue contacts is not that predicted by Crick's model. Colloquially, his model was described as "knobs into holes"; ours can be described as "ridges into grooves." Helix-sheet packing The model for packing an a-helix onto a parallel or antiparallel pleated sheet is illustrated in Fig. 3. There are two general features of pleated sheets that are important for this model: the first is the packing between neighboring residues within a sheet, and the second is its tendency to have a right-hand twist. If we consider the Ca atoms on the same side of a sheet, the distance between neighbors along the strands is 7 A and that between those in adjacent strands is 5 A. Side chain volumes vary between 25 A3 (alanine)
4132
Chemistry: Chothia et al.
Proc. Natl. Acad. Sci. USA 74 (1977)
a
N
0;..;d
/O
,0
~Q'
D
/0
0~
0-0
b
t
UI13 10.9
1.8
H
S
C
00
0
?
*
00, .'
0
00
00 L
,0f
0
06,
Q1=0
D
H
0000
0.,gO
~00,0
0
0,0
*/0
i4
4i.,'4
LD
Q>
d
C
FIG. 3. The model for helix-sheet packing. (a) The helical residues i, i + 1, i + 4, i + 5, i + 8,. . .; form a surface with a right-hand twist which is complementary to the right-hand twist of a pleated sheet shown in (b). (c) The helix is shown on top of the twisted sheet. The corners marked U are above the plane of the page and those marked D are below it. (d) Sections showing idealized helix-sheet interfaces for different values of Q (see text).
FIG. 4. Observed a-helix-pleated sheet packing. (a) Histogram of the observed Q. The values are for the 19 helices in carboxypeptidase, flavodoxin, triose phosphate isomerase, and subtilisin that have four or more residues in contact with a pleated sheet. The helix is above the sheet. (b) The normal contact residues (see text) are shown as filled circles in a flattened projection of a helix (see Fig. 1 a and b). (c) The helical residues in contact with the pleated sheet (filled circles) in flavodoxin. Twenty-three of 26 are normal contact residues. The abnormal contacts are ringed for identification.
and 170 As (tryptophan) (3). This means that most side chains in a pleated sheet will be in contact with their neighbors so the surface will not have ridges and grooves but can be considered as flat with, usually, only small irregular holes and protuberances.
The parallel and antiparallel pleated sheets found in globular proteins have a twist that is right-handed if the sheet is viewed in a direction parallel to the polypeptide chain (10). The effect of this twist is that neighboring chains wind around each other while remaining a constant distance apart. The same thing happens to the ropes in a rope ladder if a far rung is given a right-hand twist relative to a near rung. For a typical pleated sheet, the right-handed twist about an axis parallel to the chain direction is observed to be about 50 per A. Now let us consider an a-helix. The residue pairs (i, i + 1), (i + 4, i + 5), (i + 8, i + 9) wind around the helix with a right-hand twist (Fig. 3). For a regular a-helix this twist is about 6 per A. Also, given suitable side chain size and conformation, these residues can form a flattened, although irregular, surface. For reasons that will become apparent below, we shall call these
...
residues the "normal contact residues." In its simplest form, our model for a-helix-pleated sheet packing can be stated as follows: an a-helix will pack onto a pleated sheet with its axis parallel to the strands of the sheet because, in this orientation, the normal contact residues form a surface complementary to that of the sheet. Such a model would predict that the helix residues in contact with the sheet willbei,i + 1,i + 4,i + 5,i + 8,i + 9,...,andthatthe angle between the helix axis and the strands in the sheet (Q) will be O0. On a more detailed level we can see that the twist of the pleated sheet means that orientations of the helix away from
the parallel position (Q = 00) are more likely to occur in the negative (clockwise) direction (the helix is assumed to be above the sheet) (Fig. 3d). In this orientation (Q < 00), although the exposed ends of the helix move away from the sheet, its center is still able to close-pack. In the opposite orientation (Q > 00), the two ends of the helix will pack onto the sheet but they lift its center off the sheet and so create an internal cavity in the protein. Using the same argument, we can show that helices packed on sheets with a large twist will have negative Q. A dominant structural feature of the proteins flavodoxin, carboxypeptidase, subtilisin, and triose phosphate isomerase is a large central pleated sheet flanked by a-helices. We have examined the contacts that occur between the residues in these proteins and have found that they contain 19 helices that have four or more residues in contact with the face of a pleated sheet. In total, the 19 helices have 129 residues in contact with the pleated sheets, of which 112 (87%) are what we defined above as "normal contact residues" for sheet-helix packing. In Fig. 4 we show, for flavodoxin, the helical residues that are in contact with the central pleated sheet and the distribution of the values of Q. All these angles are in reasonable agreement with our model. Thirteen have values in the range -10 i 100 and the distribution is skewed toward negative values. Sheet-sheet packing We consider in this section the face-to-face packing of two pleated sheets. Inspection of protein structures shows that we can distinguish two classes of double-sheet structures: in the first class they are formed by the packing of two large and essentially independent pleated sheets; in the second class they are formed by the folding over of single sheets. In all these proteins the sheet has a right-hand twist when
Chemistry: Chothia et al.
Proc. Natl. Acad. Sci. USA 74 (1977)
4133.
(iii)
FIG. 5. The packing of two independent pleated sheets. (a) Two pleated sheets are represented by two thin smooth sheets. Arrows indicate the chain direction. Each sheet has a right-handed twist, so the corners are either above the plane of the paper (U or UU), or below the plane (D or DD). The sheets are placed face-to-face. Two sheets with the same twist close-pack when their strands are parallel. To close-pack two sheets with different twists, one sheet is rotated in a negative direction relative to the other. (b) The Ca atoms of the double-pleated sheet structures in the immunoglobulin fragment VREI (i), prealbumin (ii), superoxide dismutase (iii), and concanavalin A (iv). Filled circles denote atoms in the upper sheet and open circles those in the lower sheet. Note that the top sheet of concanavalin A is bent so that the bottom left-hand corner points down perpendicular to the plane of the page.
viewed along the strands. The extent of this twist varies from sheet to sheet. Thus, the general problem here is how sheets of different twist and residue composition associate so that the central part of the contact surface is close-packed. We shall first discuss the packing of two independent pleated sheets (Fig. 5). Let two sheets have the same degree of twist and the same residue composition. It is obvious that these close-pack when face to face with the strands in one sheet parallel to those in the other sheet (Fig. 5). Now, let one sheet have a greater twist than the other. This difference in twist can be due to a real difference in the main chain torsion angles of the two sheets or just be the surface effect of a difference in their residue compositions, or both. If these two sheets are placed face to face with their strands parallel, only two opposite corners will form contacts. Rotation of the top sheet in a clockwise (negative) direction will allow the centers of the sheets to pack more closely (Fig. 5). A rotation in the opposite (anticlockwise) direction will force the centers of the sheets to be even further apart. Computer model building shows that if T, and T2 are the twists of the two smooth sheets, the rotation angle that gives the best packing, Qc, is given by the expression:
U. For the immunoglobulin fragment VREI, QC = -32 and Q -350; for prealbumin, Qc = -36 and Q = -350; for concanavalin A, QC = -82 and Q = -300; and for superoxide dismutase, QIc = -52 and QI = -50 (Table 2). The failure of the relationship for concanavalin A is due to the pleated sheets of this protein being bent as well as twisted (Fig. 5). This relative rotation of two rectangular sheets of different twist means that their corners either lift off the packed interface or stick out at the side (Fig. 5). In real structures the corners that lift off the interface are sometimes removed by shortening the sheet at these corners. This gives the sheets a rhomboidal rather than rectangular shape and makes the double-sheet structure appear elliptical when viewed from the side in certain directions (for example, see figure 3 in ref. 11). The strands that stick out at the side sometimes fold under their own sheet and hydrogen bond to become part of the second 'sheet. There is one example
=
Table 2. Twist and relative orientation of packed pleated sheets
Twist of pleated sheets* Upper (TI) Lower (T2)
580
420
Protein Bence-Jones protein VREI Prealbumin
Relative orientations Q Qct
-320 -350
QC= -2(I7T1 T21)
-
[1]
The immunoglobulin domains, concanavalin A, prealbumin, and superoxide dismutase contain two large antiparallel pleated sheets of different twist packed face to face. A projection of these double-sheet structures is given in Fig. 5. We have measured the overall twist of each of the sheets and, using Eq. 1, have calculated the expected relative rotation, Qc Using the atomic coordinates, we have also measured the actual rotation,
640
370 290
460
630
700
-360
-520 -820
-350
-500
-300
Superoxide
dismutase
Concanavalin A
* The upper and lower sheets are defined by Fig. 5. t Defined in the text and in Fig. 5. 9,c = -2(1T1 - T21). The observed value of Q.
4134
Chemistry: Chothia et al.
b
1, D,' v.t
'U
Proc. Nati. Acad. Sci. USA 74 (1977)
14
-~f
U ,'
42
TWIST
AND
BEND
FIG. 6. The packing of pleated sheets with a right-handed supertwist. (a) Residues 14-42 of elastase are represented by open circles. These residues form a triple-stranded pleated sheet whose hydrogen bonds are shown as dashed lines ( ---- ) in i. The main chain conformation of this sheet is shown in ii; note its right-handed twist and supertwist. (b) Two pleated sheets are schematically treated as two thin smooth sheets. The strand direction in each is indicated by an arrow. They are placed face to face with their strand directions at right angle. Each sheet has a right-handed twist so that corners marked U are above the plane and those marked D are below. Note that the upper left and lower right corners are in contact while the other two splay apart. (c) The double-pleated sheet structure of first domain in elastase. The Ca atoms of each sheet residue are indicated by a circle. Filled circles show those in the upper sheet and open circles, those in the lower. Note that the two sheets are covalently joined at the upper left and lower right corners. Where the sheets splay apart, residues 43-51 and 103 are inserted to maintain the packing. Compare with b.
double sheet is formed by folding over a single sheet, a local right-handed supertwist occurs at one or both of these two corners (compare Fig. 6 b and c). This covalent link is shorter than a van der Waals contact and it pulls the two halves of the sheet together so that they are actually in contact along the diagonal joining the two close corners. As we move away from this diagonal, the twist of the sheets makes them splay apart and we often find that a short piece of polypeptide is inserted to maintain the packing (Fig. 6c). Double-sheet structures joined at two corners, as in elastase (Fig. 6c), are found in the trypsin family, staphylococcal nuclease, triose phosphate isomerase, and thermolysin. Doublesheet structures joined at one corner only occur in alcohol dehydrogenase, ribonuclease, and papain. Conclusion The packing models we have presented derive from the general geometrical properties of a-helices and pleated sheets and from the necessity to close-pack without inducing appreciable steric strain. The models implicitly ignore the exact size and shape of individual side chains. Their success shows that this is reasonable. However, our approach can be extended to include the effects of individual side chains: this might allow an exact understanding of particular packings and of those few cases that are not described by our models. In protein molecules the polypeptide chain tends to run back and forth across the molecule. Each run usually contains an a-helix or is part of a pleated sheet (12). Thus, the important intramolecular contacts that occur in a protein are between its constituent a-helices and/or its pleated sheets. Because the models that we have presented describe almost all the secondary structure packings that we have so far analyzed, the packing rules given by these models are the important determinant of the three-dimensional structure of proteins.
This paper owes much to the critical encouragement of Drs. Peter Pauling and Joel Janin. The work was initiated at the Service de Biochimie Cellulaire of the Institut Pasteur in Paris and part of it was done at the 1976 CECAM workshop on Protein Dynamics. We thank Dr. Richard Feldmann, Prof. D. C. Phillips, and Dr. C. Blake for atomic coordinates. The work was supported by grants to Dr. Peter Pauling from the Institute of Neurological and Communicative Diseases and Stroke (1 R01 NS14021-02 BBCA) and the Medical Research
Council.
1. Pauling, L., Corey, R. B. & Branson, H. R. (1951) Proc. Natl. Acad. Sci. USA 37, 205-211. 2. Pauling, L. & Corey, R. B. (1951) Proc. Natl. Acad. Sci. USA 37, 235-285. 3. Richards, F. M. (1974) J. Mol. Biol. 82, 1-14. 4. Chothia, C. H. (1975) Nature 254,304-308. 5. Ramachandran, G. N. & Sasisekharan, V. (1968) Adv. Protein Chem. 23, 284-437. 6. Richards, F. M. (1977) Adv. Biophys. Bioeng. 6, in press. 7. Gelin, B. & Karplus, M. (1975) Proc. Natl. Acad. Sci. USA 72, 2002-2006. 8. Cooper, A. (1976) Proc. Natl. Acad. Sci. USA 73,2740-2741. 9. Crick, F. H. C. (1953) Acta Crystallogr. 6,689-697. 10. Chothia, C. H. (1973) J. Mol. Biol. 75,295-302. 11. Richardson, J. S., Thomas, K. A. & Richardson, D. C. (1975) Biochem. Biophys. Res. Commun. 63,986-992. 12. Levitt, M. & Chothia, C. H. (1976) Nature 261, 552-558.
of this in the immunoglobulin fragment VREI and two examples in superoxide dismutase. Double-sheet structures that are formed by folding over a single sheet involve two related problems: how does a pleated sheet fold over, and how do the two halves pack together? Due to the combined effect of the steric limitation on possible main chain conformations (5), the right-handed twist of pleated sheets (9) and the necessity to retain interchain hydrogen bonds, pleated sheets must fold over by means of a local right-handed supertwist as illustrated in Fig. 6a. This local supertwist puts the strand directions in the two halves at approximately right angle to each other. Pleated sheets folded over by a right-handed supertwist occur in the trypsin family (Fig. 6c), staphylococcal nuclease, triose phosphate isomerase, thermolysin, alcohol dehydrogenase, ribonuclease, and papain. The way in which the two halves of the sheet pack together varies, but it is a variation on one theme. To illustrate this we use the schematic double sheet in Fig. 6b and the particular example of the first elastase domain (Fig. 6c). If the two sheets in Fig. 6b were independent of each other, they would only have two corners in van der Waals contact, the upper left and lower right in our diagram. If, however, the
Вам также может понравиться
- Journal: Structure of SpinelДокумент14 страницJournal: Structure of SpinelRaul EvertonОценок пока нет
- Chapter 6. QuasicristalesДокумент48 страницChapter 6. QuasicristalesafnaftanelОценок пока нет
- Systematics of The Spinel Structure TypeДокумент23 страницыSystematics of The Spinel Structure TypeAlin DrucОценок пока нет
- Crystallography and The Penrose Pattern: London EnglandДокумент5 страницCrystallography and The Penrose Pattern: London EnglandedirozemberghОценок пока нет
- Halpin Tsai Review ArticleДокумент9 страницHalpin Tsai Review ArticleBiju Balan LalyОценок пока нет
- Systematics of The Spinel Structure Type: Physics I) Chemistry MiheralsДокумент23 страницыSystematics of The Spinel Structure Type: Physics I) Chemistry MiheralsBassManОценок пока нет
- Yidun Wan - Effective Theory of Braid Excitations of Quantum Geometry in Terms of Feynman DiagramsДокумент23 страницыYidun Wan - Effective Theory of Braid Excitations of Quantum Geometry in Terms of Feynman DiagramsMopadDeluxeОценок пока нет
- Structure of SpinelДокумент14 страницStructure of SpinelAlin DrucОценок пока нет
- Modeling and Analysis of Discretely Supported Thin-Walled Silo Shells With Stringer Stiffeners at The SupportsДокумент8 страницModeling and Analysis of Discretely Supported Thin-Walled Silo Shells With Stringer Stiffeners at The SupportsAnonymous wWOWz9UnWОценок пока нет
- Relative Orientation of Close-Packed:, 8-Pleated Sheets in ProteinsДокумент5 страницRelative Orientation of Close-Packed:, 8-Pleated Sheets in ProteinsParijat BanerjeeОценок пока нет
- Part 8 The Triclinic SystemДокумент2 страницыPart 8 The Triclinic SystemstephenОценок пока нет
- Louis Kauffman - From Knots To Quantum Groups (And Back)Документ26 страницLouis Kauffman - From Knots To Quantum Groups (And Back)Sprite090Оценок пока нет
- The Classification of Tilted Octahedra in PerovskitesДокумент12 страницThe Classification of Tilted Octahedra in PerovskitesTran Quang Minh NhatОценок пока нет
- John E. Hearst and Nathaniel G. Hunt - Statistical Mechanical Theory For The Plectonemic DNA SupercoilДокумент7 страницJohn E. Hearst and Nathaniel G. Hunt - Statistical Mechanical Theory For The Plectonemic DNA SupercoilUylrikkОценок пока нет
- Anisotropy1,2 MBДокумент13 страницAnisotropy1,2 MBRuben AñezОценок пока нет
- Phys 203 NewДокумент103 страницыPhys 203 NewDukuzumuremyi Jean PierreОценок пока нет
- Lecture 2 - Crystalline Structure of MetalsДокумент25 страницLecture 2 - Crystalline Structure of MetalsAkerkeMami100% (1)
- The Crystal Structure: A U - STДокумент23 страницыThe Crystal Structure: A U - STMansyur SQeОценок пока нет
- Takahumi Suzuki and Naoki Kawashima - Supersolid of Hardcore Bosons On The Face Centered Cubic LatticeДокумент5 страницTakahumi Suzuki and Naoki Kawashima - Supersolid of Hardcore Bosons On The Face Centered Cubic LatticeMremefОценок пока нет
- The Structure of Planar Defects in Tilted Perovskites: RichardbeanlandДокумент21 страницаThe Structure of Planar Defects in Tilted Perovskites: RichardbeanlandTran Quang Minh NhatОценок пока нет
- Sybilla PRAДокумент12 страницSybilla PRACarlos BenavidesОценок пока нет
- Inflation Rules, Band Structure, and Localization of Electronic StatesДокумент8 страницInflation Rules, Band Structure, and Localization of Electronic StatesEduardo SaavedraОценок пока нет
- Unit - Ii Crystalstructure Analysis: - Analysis of Diffraction Patterns - Inter Planer SpacingДокумент29 страницUnit - Ii Crystalstructure Analysis: - Analysis of Diffraction Patterns - Inter Planer SpacingpariОценок пока нет
- Cuasicristales 3Документ5 страницCuasicristales 3Jackie Broglie MendozaОценок пока нет
- Nearly Free ElectronДокумент15 страницNearly Free ElectronJohnОценок пока нет
- M. P. Hertzberg, S. V. Vladimirov and N. F. Cramer - Rotational Modes of Oscillation of Rodlike Dust Grains in A PlasmaДокумент9 страницM. P. Hertzberg, S. V. Vladimirov and N. F. Cramer - Rotational Modes of Oscillation of Rodlike Dust Grains in A PlasmaDeez34PОценок пока нет
- X-Ray Diffraction: Strcuture of MaterialsДокумент55 страницX-Ray Diffraction: Strcuture of MaterialsAahil AleemОценок пока нет
- tmp9363 TMPДокумент12 страницtmp9363 TMPFrontiersОценок пока нет
- Entropy: Loop Entropy Assists Tertiary Order: Loopy Stabilization of Stacking MotifsДокумент9 страницEntropy: Loop Entropy Assists Tertiary Order: Loopy Stabilization of Stacking MotifsNepali KetoОценок пока нет
- Alain Goriely and Michael Tabor - Nonlinear Dynamics of Filaments III: Instabilities of Helical RodsДокумент20 страницAlain Goriely and Michael Tabor - Nonlinear Dynamics of Filaments III: Instabilities of Helical RodsDopameОценок пока нет
- Solid State Physics Homework Set 2 Solutions: Greg BalchinДокумент17 страницSolid State Physics Homework Set 2 Solutions: Greg BalchinAnonymous qQnnEc3Оценок пока нет
- Sundance Bilson-Thompson, Jonathan Hackett and Louis H. Kau Man - Particle Topology, Braids and Braided BeltsДокумент21 страницаSundance Bilson-Thompson, Jonathan Hackett and Louis H. Kau Man - Particle Topology, Braids and Braided BeltsMopadDeluxeОценок пока нет
- MODULE 5: Conformational Analysis of Alkanes and CyclohexanesДокумент8 страницMODULE 5: Conformational Analysis of Alkanes and CyclohexanesARMAN AKRAM BIN OMAR / UPMОценок пока нет
- Tight-Binding Calculations of The Valence Bands of Diamond and Zincblende CrystalsДокумент15 страницTight-Binding Calculations of The Valence Bands of Diamond and Zincblende CrystalsDante BaldeonОценок пока нет
- Physical Chemistry Official Material PDFДокумент248 страницPhysical Chemistry Official Material PDFMy FunОценок пока нет
- Sachin Goyal, N. C. Perkins and Christopher L. Lee - Nonlinear Dynamic Intertwining of Rods With Self-ContactДокумент35 страницSachin Goyal, N. C. Perkins and Christopher L. Lee - Nonlinear Dynamic Intertwining of Rods With Self-ContactLokosooОценок пока нет
- ArticleДокумент22 страницыArticlezrxyb7rf66Оценок пока нет
- Structure of SolidsДокумент25 страницStructure of SolidsAldi AhmadОценок пока нет
- 30residual Stresses and TheirДокумент12 страниц30residual Stresses and Theirraushan kumarОценок пока нет
- Diffuse Field Transmission Into Infinite Sandwich Composite and Laminate Composite CylindersДокумент34 страницыDiffuse Field Transmission Into Infinite Sandwich Composite and Laminate Composite Cylindersmuhammedbusra6716Оценок пока нет
- Supercoiling of Knotted and Unknotted DNA Plasmids Contact in Kirchhoff Rods With Applications To Theory of SelfДокумент20 страницSupercoiling of Knotted and Unknotted DNA Plasmids Contact in Kirchhoff Rods With Applications To Theory of SelfLokosooОценок пока нет
- Triple LinkageДокумент13 страницTriple Linkagepalerm00Оценок пока нет
- Francesca Maggioni and Renzo L. Ricca - Writhing and Coiling of Closed FilamentsДокумент16 страницFrancesca Maggioni and Renzo L. Ricca - Writhing and Coiling of Closed FilamentsVortices3443Оценок пока нет
- Fiber Diffraction PDFДокумент12 страницFiber Diffraction PDFSagar De'biomimicОценок пока нет
- Dirac Cone in Lieb LatticeДокумент29 страницDirac Cone in Lieb LatticevanalexbluesОценок пока нет
- Statistical Thermodynamics of Association Colloids: V. Critical Micelle and ShapeДокумент11 страницStatistical Thermodynamics of Association Colloids: V. Critical Micelle and ShapeAlfredo HernándezОценок пока нет
- Crystal Structure - Wikipedia, The Free EncyclopediaДокумент13 страницCrystal Structure - Wikipedia, The Free EncyclopediaPradeep ChaudhariОценок пока нет
- Earth Materials Lab 2 - Lattices and The Unit CellДокумент6 страницEarth Materials Lab 2 - Lattices and The Unit CellMukesh BohraОценок пока нет
- Laws of CrystallographyДокумент11 страницLaws of CrystallographyMalik OwaisОценок пока нет
- F. Devreux and J. P. Boucher - Solitons in Ising-Like Quantum Spin Chains in A Magnetic Field: A Second Quantization ApproachДокумент8 страницF. Devreux and J. P. Boucher - Solitons in Ising-Like Quantum Spin Chains in A Magnetic Field: A Second Quantization ApproachPo48HSDОценок пока нет
- Evidence For 2-Chain Helix in Crystalline Structure of Sodium Deoxyribonucleate Franklin Nature 1953Документ2 страницыEvidence For 2-Chain Helix in Crystalline Structure of Sodium Deoxyribonucleate Franklin Nature 1953Abhay Kumar100% (1)
- Size, Shape, and Low Energy Electronic Structure of Carbon NanotubesДокумент4 страницыSize, Shape, and Low Energy Electronic Structure of Carbon Nanotubesteju1996coolОценок пока нет
- Sum Chapter 1Документ4 страницыSum Chapter 1mdilshadshigri1000Оценок пока нет
- Solid-State Physics: Laue DiagramsДокумент6 страницSolid-State Physics: Laue DiagramspiposatОценок пока нет
- Hydrophobically-Driven Self-Assembly: A Geometric Packing AnalysisДокумент4 страницыHydrophobically-Driven Self-Assembly: A Geometric Packing AnalysisemediageОценок пока нет
- Horace T. Crogman and William G. Harter - Frame Transformation Relations For Fluxional Symmetric Rotor DimersДокумент12 страницHorace T. Crogman and William G. Harter - Frame Transformation Relations For Fluxional Symmetric Rotor DimersKiomaxОценок пока нет
- 1984 - Small - Lateral Chain Packing in Lipids and MembranesДокумент11 страниц1984 - Small - Lateral Chain Packing in Lipids and MembranesymiyazyОценок пока нет
- James H. White and Nicholas R. Cozzarelli - A Simple Topological Method For Describing Stereoisomers of DNA Catenanes and KnotsДокумент5 страницJames H. White and Nicholas R. Cozzarelli - A Simple Topological Method For Describing Stereoisomers of DNA Catenanes and KnotsLokosooОценок пока нет
- Shapes of A Suspended Curly Hair: Week Ending 14 FEBRUARY 2014Документ5 страницShapes of A Suspended Curly Hair: Week Ending 14 FEBRUARY 2014MaingotОценок пока нет
- Analysis of Microrna Transcriptome by Deep Sequencing of Small Rna Libraries of Peripheral BloodДокумент18 страницAnalysis of Microrna Transcriptome by Deep Sequencing of Small Rna Libraries of Peripheral BloodParijat BanerjeeОценок пока нет
- Toxoplasma Gondii: Infection Specifically Increases The Levels of Key Host MicrornasДокумент8 страницToxoplasma Gondii: Infection Specifically Increases The Levels of Key Host MicrornasParijat BanerjeeОценок пока нет
- Review Article: Host Cell Signalling and Mechanisms of EvasionДокумент15 страницReview Article: Host Cell Signalling and Mechanisms of EvasionParijat BanerjeeОценок пока нет
- Solvatochromic EffectДокумент17 страницSolvatochromic EffectParijat Banerjee100% (1)
- MicroДокумент11 страницMicroParijat BanerjeeОценок пока нет
- Stress ResponseДокумент16 страницStress ResponseParijat BanerjeeОценок пока нет
- Molecule of The Month: Molecular-Chameleon: Solvatochromism at Its Iridescent Best!Документ4 страницыMolecule of The Month: Molecular-Chameleon: Solvatochromism at Its Iridescent Best!Parijat Banerjee100% (1)
- Geszvain LandickДокумент12 страницGeszvain LandickParijat BanerjeeОценок пока нет
- Codon Adaptation IndexДокумент9 страницCodon Adaptation IndexParijat BanerjeeОценок пока нет
- Factors Regulating The Transcription of Eukaryotic Protein Coding Genes and Their Mechanism of Action-A ReviewДокумент14 страницFactors Regulating The Transcription of Eukaryotic Protein Coding Genes and Their Mechanism of Action-A ReviewParijat BanerjeeОценок пока нет
- Transcription FactorsДокумент32 страницыTranscription FactorsParijat BanerjeeОценок пока нет
- Molecular BiologyДокумент13 страницMolecular BiologyParijat BanerjeeОценок пока нет
- Structural Analysis of Protein Structure: Circular Dicroism Fluorescence X-Ray NMRДокумент75 страницStructural Analysis of Protein Structure: Circular Dicroism Fluorescence X-Ray NMRParijat BanerjeeОценок пока нет
- Techniques: Interpreting Chromatin Immunoprecipitation ExperimentsДокумент5 страницTechniques: Interpreting Chromatin Immunoprecipitation ExperimentsParijat BanerjeeОценок пока нет
- Symmetry Operations 42Документ43 страницыSymmetry Operations 42Parijat BanerjeeОценок пока нет
- Relative Orientation of Close-Packed:, 8-Pleated Sheets in ProteinsДокумент5 страницRelative Orientation of Close-Packed:, 8-Pleated Sheets in ProteinsParijat BanerjeeОценок пока нет
- Ch13c MotifsДокумент6 страницCh13c MotifsParijat BanerjeeОценок пока нет
- Mirkin DNA TopologyДокумент11 страницMirkin DNA TopologyParijat BanerjeeОценок пока нет
- Final RevisedДокумент36 страницFinal RevisedMaine OzawaОценок пока нет
- Question Bank IX CH-5 CELL - THE FUNDAMENTAL UNIT OF LIFEДокумент18 страницQuestion Bank IX CH-5 CELL - THE FUNDAMENTAL UNIT OF LIFENoel MammenОценок пока нет
- Residu Kanamycin HPLCДокумент2 страницыResidu Kanamycin HPLCAprelita Nurelli Dwiana100% (2)
- Chemistry 14 Biomolecules PDFДокумент19 страницChemistry 14 Biomolecules PDFinfinilifeОценок пока нет
- Protein EngineeringДокумент3 страницыProtein Engineeringkunalprabhu148Оценок пока нет
- Animal GluesДокумент12 страницAnimal GluesLucas SilveiraОценок пока нет
- Aqa A-Level Biology Cheatsheet PDFДокумент17 страницAqa A-Level Biology Cheatsheet PDFLiza Khan100% (6)
- Chapter+5 19+McKee+Suggested+ProblemsДокумент30 страницChapter+5 19+McKee+Suggested+ProblemsShivam PatelОценок пока нет
- Lab ManualДокумент10 страницLab ManualozlemОценок пока нет
- 9 Biomolecules-Notes ToДокумент6 страниц9 Biomolecules-Notes ToAshish GОценок пока нет
- Actividad 1 Tema 1Документ2 страницыActividad 1 Tema 1David BravoОценок пока нет
- 1.02 Proteins Express DNA: Gene Expression Transcription TranslationДокумент13 страниц1.02 Proteins Express DNA: Gene Expression Transcription TranslationRoberta Soto IbarrecheОценок пока нет
- List of Bachelor Thesis Projects at Theoretical PhysicsДокумент6 страницList of Bachelor Thesis Projects at Theoretical PhysicsBayu pradanaОценок пока нет
- Mark Scheme (Results) October 2019Документ24 страницыMark Scheme (Results) October 2019sammam mahdi samiОценок пока нет
- Biochemistry PINK PACOPДокумент4 страницыBiochemistry PINK PACOPEunice SofiaОценок пока нет
- 3D Structural Insights Into Glucans and Their BindingДокумент13 страниц3D Structural Insights Into Glucans and Their BindingjoseОценок пока нет
- No Animal Byproducts Conditions Skin and Hair: AC Plant Keratin PFДокумент3 страницыNo Animal Byproducts Conditions Skin and Hair: AC Plant Keratin PFAli SARIОценок пока нет
- Biology QuestionsДокумент24 страницыBiology QuestionssophiegarciaОценок пока нет
- Portfolio RdiДокумент310 страницPortfolio RdiAlj RioОценок пока нет
- Chem Sri Vagdevi AcademyДокумент6 страницChem Sri Vagdevi AcademyTammudu Abhay100% (2)
- Analytical Ultracentrifugation Techniques and Methods PDFДокумент613 страницAnalytical Ultracentrifugation Techniques and Methods PDFJesse Haney IIIОценок пока нет
- Unit 4Документ23 страницыUnit 4Jhon Rey RodeoОценок пока нет
- By Products of Fish and PrawnДокумент9 страницBy Products of Fish and PrawnNarasimha MurthyОценок пока нет
- Day 3Документ10 страницDay 3Kinberly AnnОценок пока нет
- June 2017 MSДокумент20 страницJune 2017 MSEefaОценок пока нет
- Test Bank For Biology Concepts and Investigations 4th Edition Marielle Hoefnagels 2Документ17 страницTest Bank For Biology Concepts and Investigations 4th Edition Marielle Hoefnagels 2AmberClineagwbd100% (36)
- Amino Acids & Proteins PDFДокумент135 страницAmino Acids & Proteins PDFJoelle AwarОценок пока нет
- History of EarthДокумент32 страницыHistory of Earthyanachii22Оценок пока нет
- Isolation and Characterization of DNAДокумент75 страницIsolation and Characterization of DNANathaniel CastasusОценок пока нет
- Genetics: Revised Curriculum OFДокумент44 страницыGenetics: Revised Curriculum OFAzzura Najmie Fajriyah Tamalate0% (1)