Академический Документы
Профессиональный Документы
Культура Документы
ChemDraw Training
Загружено:
Muhammad Rizki ArdiansyahАвторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
ChemDraw Training
Загружено:
Muhammad Rizki ArdiansyahАвторское право:
Доступные форматы
ChemBioDraw Ultra 12.
0 Suite
Universiti Teknologi MARA (UiTM) Faculty of Phamarcy, Puncak Alam 15th August, 2011
ChemBioDraw Ultra 12.0
August 2011
CONTENTS
INTRODUCTION TO THE CHEMBIODRAW ULTRA 12.0 SUITE ........................................... 4 CHEMBIODRAW ULTRA 12.0 GRAPHICAL USER INTERFACE (GUI) .............................................. 5 BASIC DRAWING USING THE MAIN TOOLS ................................................................................. 6 DRAWING BONDS AND ATOMS ................................................................................................................... 6 CHECKING AND CLEANING-UP STRUCTURES ............................................................................. 7 CHECK STRUCTURES..................................................................................................................................... 7 CLEAN UP STRUCTURE ................................................................................................................................ 7 STRUCT=NAME & NAME=STRUCT FUNCTIONS ......................................................................... 9 STRUCT=NAME ............................................................................................................................................. 9 NAME=STRUCT ............................................................................................................................................. 9 MORE ADVANCED DRAWING .............................................................................................................. 12 RADICALS, LONE PAIRS & CHARGES ...................................................................................................... 12 SHOWING STEREOCHEMISTRY................................................................................................................. 14 USING NICKNAMES .................................................................................................................................... 15 DRAWING TOOLS PALETTE ................................................................................................................. 17 THE ORBITAL TOOLS PALETTE ................................................................................................................ 18 GROUPING OBJECTS .................................................................................................................................. 23 THE REACTION INTERPRETER.................................................................................................................. 23 IMPORTING STRUCTURES TO CHEMBIODRAW ....................................................................... 25 CHEMBIOFINDER.COM.............................................................................................................................. 25 ISIS DRAW ................................................................................................................................................. 25 EXPORTING STRUCTURES FROM CHEMBIODRAW ................................................................. 26 EXPORTING TO CHEMBIO3D ................................................................................................................... 26 SMILES STRINGS ...................................................................................................................................... 26 EXPORTING STRUCTURES TO MICROSOFT OFFICE APPLICATIONS ................................................ 27 DRAWING AMINO ACIDS & PEPTIDES ......................................................................................... 29 AMINO ACID CODES .................................................................................................................................. 29 SEQUENCE PALETTE ................................................................................................................................... 29 VIEWING MOLECULAR ANALYSIS INFORMATION ................................................................. 32 THE FRAGMENTATION TOOLBAR ..................................................................................................... 34 THE (MASS) FRAGMENTATION TOOL ..................................................................................................... 34 THE DISSOCIATION AND RETROSYNTHESIS TOOLS ........................................................................... 35 CUSTOMISING CHEMBIODRAW (FOR PUBLICATIONS) ..................................................... 36 APPLYING DRAWING SETTINGS TO A STRUCTURE ............................................................................. 36 APPLYING PREFERENCES AND TEXT SETTINGS TO A DOCUMENT ................................................... 37 TLC PLATE TOOL ........................................................................................................................................ 38 ESTIMATING NMR CHEMICAL SHIFTS .......................................................................................... 41 PREDICT 1H NMR SHIFTS ....................................................................................................................... 41 PREDICT 13C NMR SHIFTS ...................................................................................................................... 44 INTRODUCTION TO CHEMBIO3D ULTRA 12.0 .......................................................................... 45
ChemBioDraw Ultra 12.0
August 2011
IMPORTING STRUCTURES TO CHEMBIO3D ULTRA ............................................................................. 45 THE CHEMDRAW PANEL............................................................................................................................ 45 MODEL DISPLAY ......................................................................................................................................... 46 ROTATING MODELS ................................................................................................................................. 48 X- Y- OR Z-AXIS ROTATIONS ................................................................................................................. 48 MOLECULAR COMPUTATIONS (MINIMIZING ENERGY) ...................................................... 51 CHOOSING THE BEST METHOD ................................................................................................................ 51 GAMESS MINIMIZE ENERGY COMPUTATION ..................................................................................... 53 MM2 & MMFF94 MINIMIZE ENERGY COMPUTATION ..................................................................... 55 DOCKING MODELS .................................................................................................................................... 62 FURTHER HELP ........................................................................................................................................... 64
ChemBioDraw Ultra 12.0
August 2011
Introduction to the ChemBioDraw Ultra 12.0Suite
The ChemBioDraw Ultra 12.0 suite consists of a number of individual software applications. We will concentrate on the following applications. ChemBioDraw Ultra ChemBioDraw includes Struct=Name, ChemBioDraw/Excel and ChemNMR. Create stereochemically correct structures from chemical names, and get accurate IUPAC names for structures. Estimate NMR spectra from a ChemBioDraw structure with direct atom to spectral correlation. The ChemBioDraw ActiveX/Plugin adds chemical intelligence to your browser for querying databases and displaying information. ChemBio3D Std This application for desktop molecular modeling and protein visualization provides support for state-of-the-art open GL graphics and stereo hardware. Build small molecules using the ChemBioDraw interface and see the 3D structure appear simultaneously, perform basic Molecular modeling computations such as Dihedral driver MM2 experiments, Molecular Mechanics and Dynamics.
ChemBioDraw Ultra 12.0
August 2011
ChemBioDraw Ultra 12.0Graphical User Interface (GUI)
Some of the toolbars / information windows found in ChemBioDraw:
Toolbars ChemBioDraw provides a number of optional toolbars and information windows (e.g. Chemical Properties) to facilitate production of ChemBioDraw documents. These toolbars are accessed from the View menu. They are divided into three basic groups: Standard Toolbars and Information Windows The Main Tools is an example of one of the standard toolbars. Most drawing tools (and also palettes) can be accessed through the standard toolbars. The Information Windows list chemical information about a specific molecule (or atom) which has been drawn by the user. Tool Palettes The Tool Palettes (which can also be accessed from the Main Tools) are ty pically used for more advanced drawing, such as, the drawing of chemical orbitals. Templates The Templates are essentially advanced clipart tools that allow the user to search a library of chemical (and related) drawings and molecular structures. An example
ChemBioDraw Ultra 12.0
August 2011
Basic Drawing Using the Main Tools
Drawing bonds and atoms Drawing bonds The tools for drawing bonds can all be accessed through the Main Tools. Multiple bonds are drawn using the Multiple Bonds palette. NB. To access the palette: with the cursor hovering over the palette, hold the left button of your mouse. Continue to hold the button until the required multiple bond has been selected.
Labelling atoms with the text tool The text tool is accessed through the main tools:
1. Click the Text tool. 2. Point to an atom. The pointer changes when it is positioned correctly over an atom to indicate that you will create an atom label rather than a caption. 3. Click the atom. An Atom Label text box appears over the atom.
4. Type an atom label. Numbers are automatically subscripted. 5. To close the text box, click outside the text box or select another tool.
ChemBioDraw Ultra 12.0
August 2011
Checking and Cleaning-up structures
Check Structures You can check the chemistry of a selected structure, part of a structure, or caption (in Formula style). The Check Structure command identifies errors in valences, labels and defined Nicknames. To check the valences of all selected atoms in a structure: 1. Select a structure, part of a structure, or caption with a selection tool.
2. Click the Check Structure command on the Structure menu. Each label in the structure is checked sequentially. When a label is incorrect, a message window appears.
To continue checking the structure when a message appears, click Ignore. To ignore all subsequent errors in a structure, click Ignore All. To stop checking a structure when a message appears, click Stop. Note: carbon atoms should be specifically labeled. Clean Up Structure The Clean Up Structure command is used to neaten the appearance of molecules by regularizing bond lengths and angles. Graphic objects such as arrows and aromatic circles are not affected. Use Clean Up Structure to redraw structures that you may have drawn freehand or to neaten structures that you may have imported from another application. Clean Up Structure may not create the best structure. Most structures have many "clean" forms, a problem that is particularly noticeable with straight-chain alkanes.
ChemBioDraw Ultra 12.0
August 2011
1. Select the structure or part of the structure you want to clean up with a selection tool.
2. Click the Clean Up Structure command on the Structure menu. NOTE: Clean Up Structure does not position molecules relative to other objects. Overlap may occur.
ChemBioDraw Ultra 12.0
August 2011
Struct=Name & Name=Struct Functions
Struct=Name Struct=Name generates systematic names for chemical structures with support for the Cahn-Ingold-Prelog rules for stereochemistry. Using this option, you can generate the name of structures you have drawn. Struct=Name is designed to interpret a wide variety of chemical structures. This means that you can draw a structure in the document window and, using the Convert Structure to Name command, Struct=Name will provide its name. It also automatically updates the name whenever you modify the structure. Unsupported structures Although quite useful, this feature has a few limitations for interpreting some structures such as: Sulfones and chalcogen analogs Bridged fused polycyclic structures Polymers Isotopically modified compounds Radicals, ions, and radical ions
To insert the name of a structure into your drawing: 1. Select the drawing for which you want to insert a name.
2. Click the Convert Structure to Name command on the Structure menu.
The name of the structure appears as a caption under your drawing. Name=Struct Name=Struct is a comprehensive algorithm for converting English chemical names into chemical structure diagrams. It is designed to interpret chemical names as they are actually used by chemists. In other words, it recognizes the shorthand and slang of everyday usage, in addition to recognizing most of the official IUPAC, IUBMB, and CAS rules and recommendations. In addition, it has an extensive algorithm for the identification of common typing errors, such as mehtyl. This feature helps to ensure that the generated structure is accurate.
ChemBioDraw Ultra 12.0
August 2011
Name=Struct automatically creates structures from chemical names you provide. Name=Struct recognizes most organic nomenclature. It also recognized inorganic chemistry, especially when the rules closely match those for organic chemistry. However, there area few types that are not supported: Coordination complexes Polyboranes Polymers Some highly-bridged ring systems, including fullerenes and porphyrins/porphines Some stereochemistry designators: +, -, +/-, +-, D, l, dl, endo, exo, syn, anti, r, t, c
Although some trade names are supported, Name=Struct is not intended to interpret trade or common names. A chemical database, such as chemfinder.com, is more appropriate for obtaining structures for trade or common names. Note: Because the syntax of German is very similar to that of English, Name=Struct can also interpret many German names. Converting Names to Structures There are two ways to insert a name into the document window as a structure. You can use either the Insert Structure dialog box or the Paste Special command. Insert Structure Insert Structure lets you type the name and convert it to its structure. For example, to insert 2-bromobenzoic acid: 1. Go to Structure>Convert Name to Structure. The Insert Structure dialog box appears. 2. Type 2-bromobenzoic acid. Note: You can also copy a name to the clipboard and type Crtl+V to paste the name into the dialog box. 3. If you want the name to appear below the structure, select Paste name below structure. Otherwise, the name will not be included. 4. Click OK. If you should happen to type the name incorrectly, Insert Structure will try to correct the error.Click OK to accept the suggested spelling, or click Cancel and start again to make a manual correction. Paste Special The Paste Special command allows you to paste a name, copied to the clipboard from another application, as a structure in the document window.To paste a name from the clipboard: 1. Click in the document window. 2. Go to Edit>Paste Special>Name as Structure. The structure appears in your document.
10
ChemBioDraw Ultra 12.0
August 2011
Note: If a caption is already present in the drawing area you can convert it to a structure as follows: 1. Select the caption. 2. Go to Structure>Convert Name to Structure. The molecular structure and name appear in the drawing area.
11
ChemBioDraw Ultra 12.0
August 2011
More Advanced Drawing
Radicals, lone pairs &charges
H-dot and H-dash To represent a hydrogen atom that is coming out of the plane toward you along the Zaxis, use the H-dot symbol.
To represent a hydrogen atom that is directed backwards into the plane away from you along the Z-axis, use the H-dash symbol.
You can draw H-dots and H-dashes only by clicking atoms. To draw an H-dot or an H-dash: 1. Hold down the mouse button over the Chemical Symbols tool and drag to select the symbol from the palette. 2. Click an atom.
Lone Pairs Use the lone pair symbol structure representations. To draw a lone pair: 1. Hold down the mouse button over the Chemical Symbols tool the lone pair from the palette. and drag to select to indicate a lone pair of electrons common in Lewis
2. Click the atom where you want the lone pair located, or drag it (whilst keeping the mouse left button depressed) in the appropriate direction to position it. The lone pair is offset from the atom at a fixed position.
12
ChemBioDraw Ultra 12.0
August 2011
NB. It is not possible to use the Check Structure function to check if the lone pair has been added correctly. Radicals Use the radical symbol To draw a radical symbol: 1. Hold down the mouse button over the Chemical Symbols tool the radical from the palette. 2. Point to the atom where you want the radical symbol to appear. 3. Click to draw the symbol. Radical Cation and Radical Anion Use the charge radical symbols to represent radicals that are charged. and drag to select to indicate a single non-bonded electron.
To draw a radical cation or radical anion symbol: 1. Hold down the mouse button over the Chemical Symbols tool the symbol from the palette. 2. Point to the atom where you want the symbol to appear. 3. Drag from the charge portion of the symbol to the radical portion of the symbol. Charge Symbols Use the charge symbols to represent charges on structures. To draw a charge and associate it with a structure: 1. Hold down the mouse button over the Chemical Symbols tool and drag to select the charge symbol from the palette. 2. Click to the atom to which you want the charge to correspond. The number of hydrogen atoms increases or decreases as appropriate for the addition of the charge. and drag to select
13
ChemBioDraw Ultra 12.0
August 2011
Showing Stereochemistry The bonds below indicate a single bond, down stereochemistry (into the plane of the paper, away from the viewer), from the first drawn atom to the second drawn atom.
The bonds below indicate a single bond, up stereochemistry (out of the plane of the paper, toward the viewer), from the first drawn atom to the second drawn atom.
The bond below indicates a single bond, mixture of up and down stereochemistries in some unspecified proportion.
The bonds below indicate a double bond, with cis/trans stereochemistry as drawn.
The bond below indicates a double bond, with cis/trans stereochemistry unknown.
The Show Stereochemistry calculates the absolute stereochemistry according to the Cahn-Ingold-Prelog (CIP) priority rules. For more information about the CIP rules, see Cahn-Ingold-Prelog.
14
ChemBioDraw Ultra 12.0
August 2011
Only tetrahedral and double-bond stereochemistry is supported, and only non-racemic stereochemistry is interpreted. Stereochemical indicators for aromatic bonds are not displayed. The stereochemistry feature calculates and displays the following terms: (R), (S) (r), (s) (E), (Z) Standard tetrahedral stereochemistry Tetrahedral stereochemistry determined by other stereochemical centers. For example: cis-decalin and myo-inositol. Standard double-bond stereochemistry
Stereochemistry Indicators To show the stereochemistry of the following structure:
1. Select the entire structure. 2. Right-click and click Object Settings. 3. In the Drawing tab, under Atom Indicators and Bond Indicators, select Show Stereochemistry. If you make changes to the drawing which affects the stereochemistry, the stereochemistry is recalculated. For example, select the molecule and then click Object > Flip Horizontal and the stereochemistry will alter. Using Nicknames Nicknames allow you to create short names for functional groups to use as an atom label or part of a label. When an atom is labeled with a Nickname, the chemical significance of the expanded structure is retained. Commonly used Nicknames, such as Me, Et, and Ph can be found from clicking File > List Nicknames. Generic nicknames represent a class of elements or structural moieties. The generic nicknames recognized as chemically significant by ChemBioDraw are: A: M: Q: X: R: used to represent any moiety used to represent metal used to represent a non-hydrogen heteroatom used to represent any halide undefined; used in the usual way
15
ChemBioDraw Ultra 12.0
August 2011
Defining Nicknames To define a new Nickname: 1. Create a structure containing the functional group you want to define as a Nickname. 2. Select the functional group. You must indicate the connection point for the functional group by selecting the new fragment without the bond connected to the attachment point.
NOTE: If stereochemistry is indicated in a structure that you are defining as a Nickname, the stereochemistry is retained. 3. Click the Define Nickname command on the Structure menu. The number of connection points is shown by radicals in the Formula. You can have only one or two connection points in a nickname. 4. Type a short name for the Nickname. 5. Click the OK button.
16
ChemBioDraw Ultra 12.0
August 2011
Drawing Tools Palette
The Drawing Tools palette provides shapes you can add to your drawing. These shapes are not a part of the structure they are drawn near. If you double-click on a bond, atom, or atom label with a drawing tool, they are not selected. To group drawing elements with a structure, use the Group command in the Object menu. For more information, see Grouping Objects. Some drawing element types are available with fill patterns, e.g. hollow (no fill pattern), shaded, and solid. You can change the type of box, circle, or oval using the Curves menu.
The Info window (found in the View menu) indicates the length and angle relative to the X-axis while you use any of the drawing element types. Drawing Boxes To draw a box: 1. Select one of the box tools. 2. Point where you want a corner of the box to be located. 3. Drag from one corner of the box diagonally to the opposite corner. Drawing Circles and Ovals To draw a circle or oval: 1. Select one of the circle or oval tools. 2. Point where you want the center of the circle to be located. 3. Drag outward from the center. Resizing or Rotating Drawing Elements Rectangles, circles, and ovals can now be adjusted in a similar manner to the new arrows. You no longer need to delete them and redraw from scratch. Pointing to a rectangle, circle, or oval with the same tool or a selection tool displays adjustment handles that can be dragged to reshape the object.
17
ChemBioDraw Ultra 12.0
August 2011
Circles have only a radius adjustment, but ovals and rectangles can be resized for both length and width. Rectangles also have corner handles that adjust length and width proportionately. Rectangles, circles, and ellipses can be freely rotated in three dimensions using the Structure Perspective tool:
When an object has been distorted by such rotation, dragging an adjustment point will modify the shape while preserving the perspective. Drawing Lines Lines drawn with the line tool differ in two significant ways from bonds drawn with the bond tools: They are not included in chemical interpretation of the drawing Lines that cross appear solid
To draw using a line tool: 1. Select a line tool. 2. Point where you want the line to start. 3. Drag from one end of the line to the other end. The Orbital Tools Palette The Orbital tools palette can be used to add orbitals to your chemical structures.
You draw orbitals so that the node appears first. Depending on the orbital type, available fill patterns are: Background color Shaded Solid color
18
ChemBioDraw Ultra 12.0
August 2011
You can modify the fill colors with the Color menu. To draw an orbital: 1. Select the orbital tool 2. Click the required orbital. 3. Click where you wish to draw the orbital. Display the Info window to view the orbital's length and angle relative to the X-axis while you draw it. You can constrain the length and angle of an orbital by selecting Fixed Lengths and Fixed Angles on the Object menu. .
NOTE: Orbitals are not normally part of the structure they are drawn near and are not selected when you double-click a bond, atom, or atom label with a selection tool. To group the orbitals with the structure, use the Group command from the Object menu. For more information, see Grouping Objects.
19
ChemBioDraw Ultra 12.0
August 2011
Drawing Reaction Schemes
It is possible to illustrate reactions. To demonstrate, we will draw the reaction shown below:
Drawing an arrow Starting with 2-propanone, draw the reaction arrow: 1. On the Main Tool toolbar, click the Arrow tool to display the Arrow toolbar. Note: The triangle in the lower right corner of the Arrow icon indicates it contains a palette. 2. Holding the mouse button down, move the mouse to the palette title bar, then release the mouse button. The palette becomes a floating toolbar. Drag the palette to the location you want. 3. Click the third arrow from the left in the top row. 4. After you select the arrow, close the Arrows palette. Draw the arrow you selected: 1. In the document window, click and drag the mouse horizontally to the right of the 2propanone structure. The arrow appears. 2. Select the Text tool . 3. Click above the arrow. A text box appears. 4. Type OH and press Esc. If you want to realign the text box, use a selection tool. Note: If Show Chemical Warnings is turned on (the default), a red box will appear around the OH label as soon as you change tools or open another text box indicating an error. Ignore this for now.
Add a charge symbol using the specialized symbols available in the Chemical Symbols tool palette:
1. In the Main Tool toolbar, click the Chemical Symbol tool. 2. Holding the mouse button down, select the circled negative charge symbol.
3. Point to the center of the OH label. Move the cursor slightly right or left to select the O.
20
ChemBioDraw Ultra 12.0
August 2011
4. With the oxygen atom selected, drag the charge symbol around the atom to the desired position. Objects added from the Chemical Symbols palette are associated chemically with the structure they are near. Note that the red valence error warning disappears when you add the minus charge. Curved arrows For some reactions, you may not want to be limited to drawing simple, straight arrows. In fact, you may need a variety of curved or colored arrows to enhance your drawings. You can curve most arrows found on the Arrows toolbar. After you paste an arrow in your drawing, click and drag the selection point located in the middle of the arrow As you drag the selection point, you will see the size of the arc, measured in degrees. Colored arrows You can also color straight and curved arrows. To color an arrow: 1. In the drawing window, select the arrow you want to color. 2. Select a color from the Color menu. Customizing block arrows For block arrows (those that arent made of just simple lines), you can create various colored effects using the Filled, Faded, and Shaded options. To use these effects: 1. Select a block arrow from the Arrows palette and right-click it. 2. Select a color from the Color menu. 3. On the context menu, choose either Filled, Faded, or Shaded. Try experimenting with different colors and effects to get the appearance you want. Drawing the product We now create 4-hydroxy-4-methyl-2-pentanone using a copy of the 2-propanone structure. You can also create the product from scratch but you may find copying another structure more convenient. Copy the structure:
1. Click the Marquee tool . 2. Select the 2-propanone structure and its caption.
3. Press and hold the Ctrl (windows)
The hand pointer with a plus sign indicates that you are in the duplication mode of a selection tool.
21
ChemBioDraw Ultra 12.0
August 2011
4. Drag the selection rectangle to the right and release the mouse button.
Note: When you create a copy as shown above you can drag it anywhere in the document window. If you want the copy to remain aligned with the original, hold Shift while dragging. Modify the duplicate structure:
1. Select the Solid Bond tool . 2. Click the far right bond of the copied structure.
3. Point to a terminal carbon. 4. Click the carbon atom until three bonds appear, allowing a pause between each click. Note: If you click too fast, the click is interpreted either as a double-click, which opens a text box or a triple-click, which duplicates your last atom label.
5. Point to a terminal carbon atom. 6. Type capital OH.
Replace the product caption: 1. To update the products caption, select the caption with the Text tool and type 4 hydroxy-4-methyl-2-pentanone, press the Enterkey, and type 1 mole. 2. Using the Text tool, add 2 Mole to the caption for the reactant. If the captions are not aligned properly under their structures, move them using a selection tool.
22
ChemBioDraw Ultra 12.0
August 2011
Adding a frame To complete the drawing, add a shadowed box around it:
1. Click the Drawing Elements tool . 2. Select the shadowed box from the palette of drawing elements. 3. Point to the upper left corner of the reaction scheme. Hold the down the mouse button and drag diagonally downward to the right to draw the box. Grouping Objects Some objects, such as arrows, are not associated with each other automatically. You can manually group the objects together using the Group command. You can then manipulate or move them as a single object. To group objects: 1. Click the Marquee tool 2. Do one of the following: .
Draw a box around the arrow, caption and symbol to select them. Click the arrow, then hold the shift key down while you click the OH caption and the charge symbol.
The Shift key allows you to add objects to a selection without deselecting other objects. Note: If you click a selected object while holding Shift, that object is deselected. 3. Go to Object>Group. Note: You can still select individual objects within a group by clicking them. Aligning the structures To align the structures: 1. Select both structures with the Lasso or Marquee. 2. Go to Objects>Align>T/B centers. The Reaction Interpreter After you draw your reaction, one feature you may find quite useful is the reaction interpreter. The interpreter examines your reaction and identifies its parts the reactants, products, catalysts, and annotations you may have added. The interpreter color-codes each part so that you can identify them quickly.
23
ChemBioDraw Ultra 12.0
August 2011
To use the reaction interpreter: 1. Go to View>Show Reaction Interpretation. 2. Hover your cursor over the reaction arrow. Color codes The various parts of a reaction are color-coded as follows: Redreactants Magentaproducts CyanAnything above the reaction arrow BlueAnything below the reaction arrow
Guidelines To be interpreted accurately, your reaction must follow a few guidelines: Each reaction and product must contain at least one atom. Each reaction and product must be described using a single label. Multiple reactants and/or products must be linked by a + sign. Reaction conditions must reside completely within the left and right boundaries of the reaction arrow. The reaction cannot include a curved arrow.
24
ChemBioDraw Ultra 12.0
August 2011
Importing Structures to ChemBioDraw
ChemBioFinder.Com ChemBioFinder.Com is a portal of free and subscription scientific databases. The user can search the ChemBioFinder databases by entering a chemical Name, CAS number, molecular formula or weight.
The structure can simply be copied to the clipboard (by right-clicking the structure) and pasted into a ChemBioDraw or ChemBio3D document. http://chemfinder.cambridgesoft.com/ ISIS Draw ISIS Draw is a free chemical drawing program from Symyxsomewhat similar to ChemBioDraw. Chemical structures can easily be imported from ISIS Draw to ChemBioDraw. The user may simply copy the desired structure from an open ISIS Draw file and paste it directly into a ChemBioDraw document.
A second option is to directly open an ISIS Draw sketch file in ChemBioDraw. When browsing for ISIS Draw files ensure that the Files of type box is changed to All Files (*.*) by using the drop-down arrow.
25
ChemBioDraw Ultra 12.0
August 2011
Exporting Structures from ChemBioDraw
Exporting to ChemBio3D Most commonly users will wish to export a 2D ChemBioDraw structure to ChemBio3D software. This task can be performed very easily. 1. Select a structure, part of a structure, or caption with a selection tool.
2. Click Copy on the Edit menu. 3. Open ChemBio3D and paste the structure into an open document. Alternatively, a ChemBioDraw file can simply be opened in ChemBio3D. The 2D structure is automatically converted to a 3D model. SMILES strings A SMILES string is a way to describe a chemical structure in a line of text. Several software packages use SMILES strings as a way to enter and store chemical structure information. The user may wish to convert a structure to a SMILES string in order to export the structure to another application. Creating SMILES Strings To create the SMILES string for the following structure: 1. Select the structure using a selection tool. 2. Point to the Copy As, command on the Edit menu, and choose SMILES. The SMILES string is transferred to the Clipboard.
To display the SMILES string, paste the string in a document window. When the SMILES string is on the Clipboard, you can transfer it to another application that can interpret and manipulate SMILES strings. If you select more than one structure in ChemBioDraw, and choose Copy SMILES from the Edit menu, the SMILES string for each structure is copied to the Clipboard and separated from the previous SMILES string by a period. SMILES supports an alternate notation for aromatic structures using lowercase letters. ChemBioDraw generates this type of SMILES string for any structure drawn with explicit aromatic bonds, either by using the Aromatic bond type in the Atom Properties dialog
26
ChemBioDraw Ultra 12.0
August 2011
box, or by placing a circle (from the Drawing Elements palette) within any ring structure.
c1ccccc1
C1=CC=CC=C1
Convert SMILES string to a ChemBioDraw structure If you have a SMILES string, then you can automatically convert that text string into a ChemBioDraw structure. To paste a SMILES string as a ChemBioDraw structure: 1. Select the SMILES string using the Text tool. 2. Click the Copy command on the Edit menu. 3. Point to Paste Special on the Edit menu, and choose SMILES. The SMILES string is drawn as a structure. Exporting Structures to Microsoft Office Applications ChemBioDraw structures may be easily exported to Microsoft Office applications such as Word or Excel. 1. Select a structure, part of a structure, or caption with a selection tool.
2. Click Copy on the Edit menu. 3. Open the Word or Excel document and paste the structure at the desired place. Once the structure is inserted in the document it is still possible to edit the structure in ChemBioDraw. By right-clicking the structure within the Microsoft Office document the user is given the option of opening and editing the structure within ChemBioDraw.
27
ChemBioDraw Ultra 12.0
August 2011
If the user does not wish to subsequently edit the structure it is best to use the Paste Special function within the Microsoft Office document and the paste the structure as a picture. This should eliminate any compatibility issues if a later user does not have ChemBioDraw installed on their PC.
28
ChemBioDraw Ultra 12.0
August 2011
Drawing Amino Acids& Peptides
You can draw a peptide chain by: Stringing Stringing is a bond Stringing the amino acid 3-letter codes together. the amino acid 3-letter codes together in an atom label as long as there attached. the amino acid 3-letter codes together and not including bonds.
Amino Acid Codes Amino Acid Alanine Arginine Asparagine Aspartic acid Cysteine Glutamic acid Glutamine Glycine Histidine Isoleucine 3-Letter Ala Arg Asn Asp Cys Glu Gln Gly His Ile 1-Letter A R N D C E Q G H I Amino Acid Leucine Lysine Methionine Phenylalanine Proline Serine Threonine Tryptophan Tyrosine Valine 3-Letter Leu Lys Met Phe Pro Ser Thr Trp Tyr Val 1-Letter L K M F P S T W Y V
Convert Strings of 3-Letter Codes to a ChemBioDraw Structure 1. Draw the peptide chain as described above. 2. Select the peptide chain using a selection tool. 3. Right-click the peptide and select Copy As SMILES from the Molecule menu. 4. From the Edit menu of the main toolbar select Paste Special SMILES The peptide chain may also be imported into ChemBio3D in the same manner.
Sequence Palette ChemBioDraw version 12.0also contains a feature for drawing sequences of amino acids and nucleotides.
29
ChemBioDraw Ultra 12.0
August 2011
The Main toolbar includes a unique feature that enables you to draw protein and RNA structures quickly. Using labels specifically assigned to nucleic and amino acids, you first draw a sequence. Then, you can expand, contract or remove labels to get the drawing you want. To draw a sequence: 1. On the Main Toolbar, click the Sequence icon and choose one of the following tools: Single-letter amino acid tool This tool lets you create a protein chain using a one-letter label to represent each amino acid. Three-letter amino acid tool Create a protein chain using three-letter amino acid labels. DNA tool This lets you create a DNA chain using labels that represent each of the nucleic acids. RNA tool This lets you create an RNA chain using nucleic acid labels. 2. Click in the drawing window where you want to start the sequence. A text box appears. 3. In the text box, enter the single- or three-letter label for the residue you want in the chain. Note: The IUPAC codes for all nucleotide base and amino acid labels are listed below. 4. Repeat step 3 for each residue you want. 5. When your chain is complete, click anywhere in the drawing window or choose a different tool. Expanding sequences After you create a sequence, you can expand it into its structure. If you dont want to expand the entire sequence, you can choose only those labels you want. To expand a sequence: 1. Using either the Marquee tool or Lasso tool Hold down the SHIFT key to select more than one. , select a label in the sequence.
30
ChemBioDraw Ultra 12.0
August 2011
2. Go to Structure>Expand Label. The labels you chose expand into their structures. Collapsing labels When you expand a label into its structure, the label remains on screen and appears below the structure. To collapse the structure back to its label (in the sequence): 1. Double-click the label to select the structure. 2. Go to Structure>Contract Label. Removing Residues When you remove a residue, the adjacent labels close, keeping the sequence intact. To remove a residue: 1. If the label is expanded, collapse the structure back to its label. 2. Double-click the label. 3. Press DELETE on your keyboard. Replacing Residues To replace a residue, select the Marquee tool and hover your mouse over it. Then, type the new hotkey. Adding Labels To add a new label: 1. Choose the appropriate sequencing tool. 2. Select an existing label you want to the new label to be adjacent to. 3. Type the new label. The new label is added to the right of the existing label. Note: To add a label so that it is the farthest left in the sequence, select the amino group (protein sequence) or 3 group (DNA/RNA sequence) Use the following codes to enter nucleotides in your sequence. IUPAC nucleotide code A C G T (or U) R Y S W Base Adenine Cytosine Guanine Thymine (or Uracil) A or G C or T G or C A or T IUPAC nucleotide code K M B D H V N . or Base G or T A or C C or G or T A or G or T A or C or T A or C or G any base gap
31
ChemBioDraw Ultra 12.0
August 2011
Viewing Molecular Analysis Information
The Analysis window displays the chemical formula, exact mass, molecular weight, m/z, and elemental analysis for the entire document, a structure, part of a structure, or a caption in Formula style. To view analysis information, click the Show Analysis Window command on the View menu. The Analysis window appears:
Values for selected objects in the document window are shown. If nothing is selected in your document, values for the entire document are shown. You can have this window open as you draw in the document. It shows the current values as you draw. The Decimals setting applies to Exact Mass, Molecular Weight, and m/z only. Analysis properties Formula: The molecular formula showing the exact number of atoms of each element in the molecule and charges, radicals, and isotopes. Exact Mass: The exact molecular mass of the structure, where atomic masses of each atom are based on the most common isotope for the element. Molecular Weight: The average molecular mass of the structure, where atomic masses are based on the natural abundance of all isotopes of the element. m/z: Mass/charge, where charge =1. The weights of the most common isotopes and a graphical representation of the isotopic abundance is shown.The molecular weight shown takes the isotopes for each atom and their natural abundance into account. Where there is more than one abundant isotope, ChemBioDraw computes multiple molecular weights. Low abundance combinations (whether because the isotope is in low abundance or because it includes many moderate-abundance contributions) are not taken into account. Elemental Analysis: The percent by weight of each element in the structure.
32
ChemBioDraw Ultra 12.0
August 2011
The information in the label updates automatically as you edit the structure. You can show or hide the information as follows: 1. Right-click in the label. It does not matter which tool you are using. 2. Point to Analysis, and select or deselect the item you want to show/hide. You can click Paste within the Analysis Window to paste the details from this window into the document.
33
ChemBioDraw Ultra 12.0
August 2011
The Fragmentation Toolbar
Features three functions: 1. Fragmentation tool splits molecules across specific bonds. 2. Dissociation tool breaks bonds and draws a reaction. 3. Retrosynthesis tool breaks bonds and draws a reaction.
The (Mass) Fragmentation tool This allows you to break existing structures across one or more bonds. This mimics the molecular fragmentation in a mass spectrometer, but does not have any predictive qualities: you must specify what bonds are to be broken. To fragment a structure, drag the mouse across one or more bonds. A line is drawn across the bonds that disappears when you release the mouse button. The operation is not final until the mouse is released. This means you can drag the line around holding the mouse button down to see different possibilities, and still back out without breaking any bonds. When the mouse button is released, any bonds crossed by the line are broken
If you want the line to remain on display, hold down the Alt key after drawing the line.
NOTE: The order in which you release the mouse button and the Alt key determines whether the line remains displayed or not. Releasing the mouse button first leaves the line displayed. The Alt key also allows you to "turn corners" and cut more than one bond.
34
ChemBioDraw Ultra 12.0
August 2011
When the line crosses a bond, the formula and exact mass for the fragments on either side of the bond are displayed as if the bond were homolytically broken. That is, a single bond turns into a monoradical on each fragment; a double bonds turns into a pair of diradicals. If more than one bond is crossed, all fragments on each side of the line are considered together. If the only bond crossed is a ring bond, a single formula/mass pair is displayed. The Dissociation and Retrosynthesis tools These two tools speed the drawing of synthesis reactions. Both tools are used in a similar manner to the Mass Fragmentation tool: they are used to cut a bond or bonds in a product. The Dissociation tool draws the resulting fragments as a standard reaction. The Retrosynthesis tool draws the reaction with the product on the left and a broad arrow pointing to the reactants, as is customary in retrosynthetic analyses. In most cases, you will have to clean up the reaction by adding or deleting atoms or bonds.
35
ChemBioDraw Ultra 12.0
August 2011
Customising ChemBioDraw (for publications)
Example journal: Drug Metabolism Letters Chemical Structures: Chemical structures must be prepared according to the guidelines below. Structures should be prepared using a suitable drawing program e.g. ChemBioDraw and provided as separate file, submitted both on disk and in printed formats.Numbers in bold should identify all chemical structure. If a name is also used to describe the compound, the associated number in bold should be in parenthesis, e.g. compound (1). Structure Drawing Preferences: [as according to the ACS style sheet] Drawing Settings: Chain angle Bond spacing Fixed length Bold width Line width Margin width Harsh spacing Text Settings: Font Size 120 18% of width 14.4 pt (0.500cm, 0.2in) 2.0 pt (0.071cm, 0.0278in) 0.6 pt (0.021cm, 0.0084in) 1.6 pt (0.096cm) 2.5 pt (0.088cm, 0.0347in) Times New Roman / Helvetica 8 pt
Under the Preference Choose: Units points Tolerances 3 pixels Under Page Setup Use: Paper US letter Scale 00%
Applying Drawing Settings to a Structure To apply these specified settings to a structure: 1. Using either the Marquee tool or Lasso tool , select a structure.
2. Go to Object>Object Settings. The following window should open:
36
ChemBioDraw Ultra 12.0
August 2011
Type the specified settings in the relevant boxes. Applying Preferences and Text Settings to a Document The Preferences box is accessed through the File menu whilst the text settings can be altered directly via the Text menu.
37
ChemBioDraw Ultra 12.0
August 2011
TLC Plate Tool
The TLC Tool is a drawing tool that allows you to easily depict Thin Layer Chromatography plates within a ChemBioDraw document. The tool creates a rectangular plate with origin line, solvent front, and one or more lanes. The lanes can be populated with spots of different Rf, size, shape, or color. To create a TLC plate: 1. Select the TLC tool from the Main Tools.
The number of lanes is a function of the width of the plate you create.
2. Drag in any direction from the point of origin.
You can manipulate parts of the plate as follows:
If you want to... Change the height or width of the plate.
then... Drag a border or corner. The number of lanes will not change.
Move the origin or solvent front. Drag the origin or solvent front line. 1. Right-click in the plate. Show or hide the origin, solvent front, borders, or side ticks. 2. Select the appropriate action.
38
ChemBioDraw Ultra 12.0
August 2011
NOTE: Use the Transparent option if you want to overlay the TLC plate on a scanned plate. Change the order of lanes. Drag the origin tick to the new location. (The lanes will remain evenly distributed along the origin.)
Add, delete, or duplicate a lane. 1. Position the cursor at the origin of the lane you want to delete or duplicate (or between lanes to add). 2. Right-click in the plate. 3. Select the appropriate action. NOTE: You can also delete lanes with the Eraser tool. Move a spot. Duplicate a spot. Delete a spot. Drag the spot. The Rf displays as you drag. Ctrl+drag the spot. Click on the spot with the Eraser tool. NOTE: If you erase all spots in a lane, the lane will be deleted.
Display or set the Rf for a spot;1. Right-click on a spot. 2. Point to TLC Spots, and take the appropriate action. add a custom spot NOTE: To display Rf for all spots, Right-click in the plate and select Show Rf from the TLC Spots submenu. Change the style or color of a 3. Right-click on a spot. spot. 4. Choose the style or color. NOTE: To change the style for all spots, Right-click in the plate and choose the style from the TLC Spots submenu. Enlarge or "smear" a spot. Shift+drag the spot. When you position the cursor on a spot and press "Shift", the cursor assumes one of three shapes, depending on how it is positioned: 1. Cross arrows 2. Horizontal arrow 3. Vertical arrow used to enlarge a spot. used to widen a spot. used to elongate a spot or create a crescent.
NOTE: You may want to zoom in (by using the F7 and F8 keys) to make positioning easier.
More on Resizing & Reshaping Spots You can resize spots by holding down the Shift key while pointing at an edge of the spot. Dragging the left or right sides of the spot will adjust the spot's width only. Dragging from the top will adjust its height only. Dragging from the bottom will adjust the tail only. Dragging from any of the other four corners will scale all three values. By using combinations of these dragging motions, you can reproduce any sized or shape spot you want.
39
ChemBioDraw Ultra 12.0
August 2011
Custom Spots The TLC tool can create the most common spot shapes. Other types of spots, such as smears, can be reproduced as custom spots. The Add Custom Spot command allows you to insert a graphic file for a spot. Typically, these files would be produced by scanning a TLC plate and saving the spots in a library of spot shapes. How to add a custom spot 1. Right click the spot that you wish to change.
2. Select the graphics file that you wish to use.
40
ChemBioDraw Ultra 12.0
August 2011
Estimating NMR Chemical Shifts
ChemNMR is a feature of ChemBioDraw Ultra only. ChemNMR estimates and displays proton and carbon-13 chemical shifts in ppm for a selected molecule. Based on 4000 parameters for 13C NMR and almost 3000 parameters for 1H NMR, chemical shifts are predicted using additivity rules and several strategies of approximation. Stereochemistry of ethylenes (cis/trans) and cyclohexanes (eq/ax) are also implemented. In case of 13C NMR, over 95 % of the shifts can be predicted with a mean deviation of 3.8 ppm. In case of 1H NMR, shifts of about 90 % of all CHx-groups can be predicted with a mean deviation of 0.2 - 0.3 ppm. But the use of polar solvents may strongly increase these deviations. 1H NMR shifts of hydrogen atoms bonded to hetero atoms are not predicted because they are affected by solvents, concentration, impurities and steric effects. Predict 1H NMR Shifts To view 1H NMR information: 1. Select the target chemical structure using the select tools.
2. Click the 1H-NMR Shifts command on the Structure menu. ChemNMR redraws the molecule with the estimated shifts and displays the information and line spectrum in a new window as shown below.
41
ChemBioDraw Ultra 12.0
August 2011
To view the spectral assignments: 1. Click the Lasso or Marquee tool. 2. Place the pointer over a peak. The assigned atoms or bonds are highlighted.
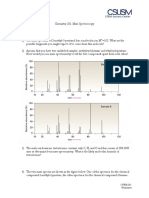
Example 1H NMR Spectra
42
ChemBioDraw Ultra 12.0
August 2011
1,1,2-Trichloroethane
1
H NMR spectrum
(Varian catalogue, spectrum no. 2, reproduced from Spectroscopic Methods in Organic Chemistry, 5th Ed., 1995, Williams & Fleming)
Ethane
1
H NMR spectrum
(Varian catalogue, spectrum no. 14, reproduced from Spectroscopic Methods in Organic Chemistry, 5th Ed., 1995, Williams & Fleming)
43
ChemBioDraw Ultra 12.0
August 2011
1-Nitropropane
1
H NMR spectrum
(Varian catalogue, spectrum no. 42, reproduced from Spectroscopic Methods in Organic Chemistry, 5th Ed., 1995, Williams & Fleming)
Predict To view
13
C NMR Shifts
13
C NMR information:
1. Select the target chemical structure using the select tools.
2. Click the 13C-NMR Shifts command on the Structure menu.
44
ChemBioDraw Ultra 12.0
August 2011
Introduction to ChemBio3DUltra12.0
Importing structures to ChemBio3DUltra The simplest way of viewing a molecule in ChemBio3D is to open a ChemBioDraw file. 1. Click on the file menu.
2. In the dialogue box that opens browse for ChemBioDraw files. The 2D ChemBioDraw file is automatically converted to a 3D ChemBio3D file on opening. The ChemDraw Panel ChemBio3Dmakes it easier than ever to create or edit models in ChemBioDraw. The ChemDraw panel is activated from the View menu. By default it opens on the right side of the screen, but like the toolbars you can have it "float" or attach it anywhere you like. The easiest way to draw a molecule in the ChemDraw panel is by using the Main Tools. If the Main Tools are not visible right-click somewhere in the ChemDraw panel and select Show Main Tools from the menu. The molecule can then be drawn within the ChemDraw Panel in the same manner as if ChemBioDraw software was being used. As the 2D structure is drawn it will appear as a 3D structure on the main screen.
45
ChemBioDraw Ultra 12.0
August 2011
Model Display The Model Display feature (accessed through the view menu) allows the user to customize the appearance of the 3D model. Structure Displays Structures are graphical representations based on the traditional physical threedimensional molecular model types. The following structure display types are available from Model Display view of the Chem3D Setting dialog box: Wire Frame Sticks Ball and Stick Cylindrical Bonds Space Filling Ribbons Cartoons
To change the default structural display type of a model: 1. From the File menu, choose Model Settings. The Chem3D Setting dialog box appears. 2. Select the Model Display tab. The Model Display control panel of the Chem3D Setting dialog box appears. 3. Set the new options.
To change the structural display type of a model temporarily: 1. Click the arrow on the Model Display tool display type. , and select the
46
ChemBioDraw Ultra 12.0
August 2011
Ribbon / Cartoon Models Ribbon and Cartoon models show large protein molecules in a form that highlights secondary and tertiary structure. Ribbon models can be colored by Group to help identify the amino acid constituents. Your model must have a protein backbone in order to display ribbons. The following caveats apply to the Ribbon and Cartoon model display types: They do not provide pop-up information. They should be printed as bitmaps.
47
ChemBioDraw Ultra 12.0
August 2011
Rotating Models
ChemBio3D allows you to freely rotate the model around axes. When you select the Trackball tool, four pop-up rotation bars are displayed on the periphery of the model window. You can use these rotation bars to view your model from different angles by rotating around different axes. You can also open the Rotate dialog box where you can use the rotate dial or type the number of degrees to rotate. To display the Rotation bars, select the Trackball tool from the Building toolbar.
When you mouse over an edge of the model window, the Rotation bars appear on the edges of the Model window. X- Y- or Z-Axis Rotations To rotate a model about the X-, Y-, or Z-axis: 1. Point to the appropriate Rotation bar. 2. Drag the pointer along the Rotation bar. If Show Mouse Rotation Zones is selected on the GUI (graphical user interface) tab of the Preferences dialog box, the rotation bars will pop up. This is the default setting. Note: The rotation bars are active when the Trackball tool is selected, even if they are hidden. The number of degrees of rotation appears in the Status bar (at the bottom of the screen). Rotating Fragments If more than one model (fragment) is in the model window, you can rotate a single fragment or rotate all fragments in the model window. To rotate only one fragment: 1. Select an atom in the fragment you want to rotate. 2. Hold the Shift key while dragging a rotation bar. To rotate all fragments, drag a rotation bar. Rotating Around a Bond To rotate the model around a specific bond: 1. Select a bond. 2. Drag the pointer along the Rotate About Bond Rotation bar on the left side of the Model window. Rotating Around a Specific Axis You can rotate your model around an axis you specify by selecting any two atoms in your model. You can add dummy atoms (see Undefined Bonds and Atoms) as fragments to specify an axis around which to rotate. To rotate the model around an axis: 1. Select any two atoms. 2. Drag the pointer along the Rotate About Bond Rotation bar on the left side of the Model window.
48
ChemBioDraw Ultra 12.0
August 2011
Trackball Tool Use the Trackball tool to freely rotate a model. Starting anywhere in the model window drag the pointer in any direction The Status bar displays the X and Y axis rotation. Internal Rotations Internal rotations alter a dihedral angle and create another conformation of your model. You can rotate an internal angle using the dihedral rotators on the Rotation Dial.
Using the Rotation Dial The Rotation Dial offers a quick method for rotating a model or dihedral a chosen number of degrees with reasonable accuracy. To open the rotation dial: 1. Click the arrow next to the Trackball tool. 2. Enter exact numbers into the degree text box. The Internal Rotation icons are available only when atoms or bonds have been selected in the model.To perform internal rotations in a model, you must select at least two atoms or one bond. You may then either rotate the model around the bond axis, or rotate either end. Note: Using the Internal Rotation Bar (the one to the left of the workspace) rotates the model around the selected bond axis. Note that this is a change from earlier versions of ChemBio3D, where the Internal Rotation Bar rotated the dihedral. Internal rotation is typically specified by a bond. The fragment at one end of the bond is stationary while the fragment attached to the other end rotates. The order in which you select the atoms determines which fragment rotates. For example, consider ethoxybenzene (phenetole). To rotate about the C-O bond where the phenyl group moves: 1. Click the arrow next to the Trackball tool, and drag the Rotation Dial onto the workspace. 2. Hold down the S key, and select the O atom. 3. Hold down the Shift and S keys, and select the C1 atom. 4. Click the left-hand dihedral rotator . 5. Drag the dial to rotate the phenyl group.
49
ChemBioDraw Ultra 12.0
August 2011
To perform a rotation about the C-O bond where the ethyl group moves, do one of the following: 1. Click the right-hand dihedral rotator. 2. Reverse the order of selection: first select C1, then O. Tip: To deselect the atoms, hold down the S key and click anywhere in the model window. Rotating a Dihedral Angle You can select a specific dihedral angle to rotate. To rotate a dihedral: 1. Select four atoms that define the dihedral. 2. Select one of the dihedral rotators on the Rotation Dial. 3. Drag the dial or enter a number in the text box. Tip: The keyboard shortcuts for dihedral rotation are Shift+B and Shift+N. You can change the orientation of your model along a specific axis. Although your model moves, the origin of the model (0, 0, 0) does not change, and is always located in the center of the model window. To change the origin, see Centering a Selection.
50
ChemBioDraw Ultra 12.0
August 2011
Molecular Computations (Minimizing Energy)
Methods Overview Minimizing energy is generally the first molecular computation performed on a model. Computational chemistry encompasses a variety of mathematical methods that fall into two broad categories: 1. Molecular mechanics Applies the laws of classical physics to the atoms in a molecule without explicit consideration of electrons. ChemBio3D 12.0 provides the following methods: Molecular mechanical methods: MM2 (directly). MM3 and MM3-protein through the ChemBio3D Tinker interface.
2. Quantum mechanics Relies on the Schrdinger equation to describe a molecule with explicit treatment of electronic structure. Quantum mechanical methods can be subdivided into two classes: ab initio and semiempirical. ChemBio3D12.0 provides the following methods: Semiempirical Extended Hckel, MINDO/3, MNDO, MNDO-d, AM1 and PM3 methods through ChemBio3D and CS MOPAC or Gaussian. Ab initio methods through the ChemBio3D Gaussian or GAMESS interface.
Choosing the Best Method Not all types of calculations are possible for all methods and no one method is best for all purposes. For any given application, each method poses advantages and disadvantages. The choice of method depend on a number of factors, including: The nature and size of the molecule The type of information sought The availability of applicable experimentally determined parameters (as required by some methods) Computer resources
The three most important of the these criteria are: Model size The size of a model can be a limiting factor for a particular method. The limiting number of atoms in a molecule increases by approximately one order of magnitude between method classes from ab initio to molecular mechanics. ab initio is limited to tens of atoms, semiempirical to hundreds, and molecular mechanics to thousands. Parameter Availability Some methods depend on experimentally determined parameters to perform computations. If the model contains atoms for which the parameters of a particular method have not been derived, that method may produce invalid predictions. Molecular mechanics, for example, relies on parameters to define a force-field. A force-field is only applicable to the limited class of molecules for which it is parametrized. Computer resources Requirements increase relative to the size of the model for each of the methods.
51
ChemBioDraw Ultra 12.0
August 2011
ab initio: The time required for performing calculations increases on the order of N 4, where N is the number of atoms in the model. Semiempirical: The time required for computation increases as N 3 or N2, where N is the number of atoms in the model. MM2: The time required for performing computations increases as N 2, where N is the number of atoms. In general, molecular mechanical methods require less computer resources than quantum mechanical methods. The suitability of each general method for particular applications can be summarized as follows. Method Type Molecular Mechanics Advantages Disadvantages Particular force field applicable only for a limited class of molecules Best For
Least intensive computationally. Gaussian is fast and is (e.g. MM2, MMFF94) useful with limited computer resources. It can be used for Uses classical physics and relies on molecules as large as enzymes. force-field with embedded empirical parameters
Large systems that consist of thousands of atoms and Systems or processes with no Does not calculate breaking or forming of bonds electronic properties Requires experimental data (or data from ab initio) for parameters Requires experimental data (or data from ab initio) for parameters Less rigorous than ab initio methods Medium-sized systems that consist of hundreds of atoms. Also, systems involving electronic transitions.
Semiempirical (e.g. MOPAC) These use quantum physics, experimentally derived empirical parameters, and extensive approximation.
Less demanding computationally than ab initio methods Capable of calculating transition states and excited states
ab initio (e.g. GAMESS)
Useful for a broad range Computationally of systems intensive Does not depend on experimental data
These use quantum physics, are rigourously Capable of calculating mathematical transition states and methods, and use no excited states empirical parameters
Small systems that consist of only tens of atoms or systems involving electronic transitions. Molecules or systems without available experimental data (new chemistry). Systems requiring rigorous accuracy.
52
ChemBioDraw Ultra 12.0
August 2011
GAMESS Minimize Energy Computation To perform a GAMESS Minimize Energy computation on a model:
1. Go to Calculations>GAMESS>Minimize (Energy/Geometry). The Minimize Energy dialog box (GAMESS Interface) appears with the Job & Theory tab displayed. 2. Use the tabs to customize your computation. See the following sections for details. 3. Click Run.
53
ChemBioDraw Ultra 12.0
August 2011
The Job & Theory Tab Use the Job & Theory tab to specify the combination of basis set and particular electronic structure theory. By default, this tab is optimized for setting up ab initio computations. For more detailed information, see the $BASIS section of the GAMESS documentation. To specify the calculation settings: 1. From the Method list, choose a method. 2. From the Wave Function list, choose a function. 3. From the Basis Set list, choose the basis set. Note: To use a Method or Basis Set that is not on the list, type it in the Additional Keywords section on the General tab. For more information, see Specifying the General Settings. 4. From the Diffuse list, select the diffuse function to add to the basis set. 5. Set the Polarization functions. If you select a function for Heavy Atom, also select an H option. 6. Select a Spin Multiplicity value between 1 and 10.
54
ChemBioDraw Ultra 12.0
August 2011
MM2 & MMFF94 Minimize Energy Computation ChemBio3D offers two commonly accepted force field calculation methods, MM2 and MMFF94. These methods are designed so that you can calculate molecular properties of your models. Each of these methods enable you to calculate a variety of steric energy, thermal energy, and other values. Results are saved as part of the atom properties. MM2 and MMFF94 may be viewed as different calculation techniques you use to arrive at a specified result. Which technique you use depends on your type of model and the property you want to calculate. For example, for a given model, you may find that one method provides a better potential energy prediction than the other. Meanwhile, the other method may produce charge values that are closer to experimental values. Here we describe each of these methods and the calculations you can perform using them. MM2 The MM2 force field method is available in all versions of ChemBio3D. MM2 is most commonly used for calculating properties of organic molecular models. The MM2 procedures described assume that you understand how the potential energy surface relates to conformations of your model. If you are not familiar with these concepts, see MM2 Note: In ChemBio3D 12.0, the MM2 atom type is used for force field calculations. In earlier versions, MM2 was used only in building models. MMFF94 Use MMFF94 to perform energy minimization calculations on proteins and other biological structures. The MMFF94 force field is available in ChemBio3D Ultra. Displaying MMFF94 Atom Types You can display the MMFF94 atom types for your model without performing calculations. The Atom Property table lists each atom, its atom type, and its charge. To view the list of MMFF94 atom types in your model: 1. Go to View>Atom Property Table. 2. Go to Calculations>MMFF94>Set Up MMFF94 Atom Types and Charges. Calculating potential energy You can perform an MMFF94 calculation of the potential energy for your model. You dont need to perform an energy minimization beforehand. 1. Go to Calculations>MMFF94>Calculate MMFF94 Energy and Gradient. 2. Go to View>Atom Property Table to view the results. Energy Minimization using MMFF94 To perform an MMFF94 minimization: 1. Go to Calculations>MMFF94>Do MMFF94 Minimization 2. In the Do MMFF94 Minimization dialog box, select any of the following options as desired and click Run when finished: Display Every Iteration Select this option to view the model during the calculation process. Keep in mind that displaying or recording each iteration may significantly add to the time required to minimize the structure.
55
ChemBioDraw Ultra 12.0
August 2011
Copy Measurements to Output Box View the value of each measurement in the Output window. Setup new Atom Types before Calculation When this option is selected, ChemBio3D will delete any custom MMFF94 atom types you have defined for your model. Deselect this option if you want to keep them. Setup new Atom Charges before Calculation When this option is selected, ChemBio3D 12.0 will replace any custom charges you have entered in the Atom Property table. If you want to retain the charges you have entered, deselect this option. Energy Minimization using MM2 To minimize the energy of the molecule based on MM2 Force Field: 1. Build the model for which you want to minimize the energy. 2. To impose constraints on model measurements, set Optimal column measurements in the Measurement table (go to View>Measurement table). 3. Go to Calculations>MM2> Minimize Energy. The Minimize Energy dialog box appears. 4. In the Minimization Energy dialog box, select any of the following options as desired and click Run when finished: Minimum RMS Gradient Specify the convergence criteria for the gradient of the potential energy surface. Use large values for shorter calculation times but less accurate results. Use a smaller value for more accurate results but longer calculation time. The default value of 0.100 is a reasonable compromise between accuracy and speed. Display Every Iteration Select this option to view the model during the calculation process. Keep in mind that displaying or recording each iteration may significantly add to the time required to minimize the structure. Copy Measurements to Output Box View the value of each measurement in the Output window. Select Move Only Selected Atoms Restrict movement of a selected part of a model during the minimization. Constraints are not imposed on any term in the calculation and the results are not affected. Note: If you plan to make changes to any of the MM2 constants, such as cutoff values or other parameters used in the MM2 force field, first make a backup copy of the parameter tables. This will ensure that you can get back the values that are shipped with ChemBio3D, in case you need them. Note: ChemBio3D 12.0 guesses parameters if you try to minimize a structure containing atom types not supported by MM2 (such as inorganic complexes where known parameters are limited). You can view all parameters used in the analysis using the Show Used Parameters command. See Showing Used Parameters. Iteration data appears in the Output window when the minimization calculations begin. The data is updated for every iteration of the computation, showing the iteration number, the steric energy value at that iteration, and the RMS gradient. (However, if
56
ChemBioDraw Ultra 12.0
August 2011
you have not selected the Copy Measurements to Output option, only the last iteration is displayed). After the RMS gradient is reduced to less than the requested value, the minimization ends, and the final steric energy components and total appear in the Output window. Intermediate status messages may appear in the Output window. A message appears if the minimization terminates abnormally (usually caused by a poor starting conformation). To interrupt a minimization that is in progress, click Stop in the Computing dialog box:
Multiple Calculations If you want, you can minimize several models simultaneously. If a computation is in progress when you begin minimizing a second model, the minimization of the second model is delayed until the first one has finished. If you are using other applications, you can run minimization with ChemBio3D in the background. You can perform any action in ChemBio3D that does not move, add, or delete any part of the model. For example, you can move windows around during minimization, change settings, or scale your model. Example: Minimizing Ethane Ethane is a simple example of minimization, because it has only one minimum-energy (staggered) and one maximum-energy (eclipsed) conformation. To minimize energy in ethane: 1. Go to View>Model Display>Display Mode>Ball & Stick. 2. Draw a model of ethane in an empty window. 3. Go to View>Model Display>Show Serial Numbers. 4. Go to Calculations>MM2>Minimize Energy. 5. Click Run in the Minimize Energy dialog box. The results of the calculation appear in the Output box. If necessary, use the scroll bar to view all the results. The total steric energy for the conformation is 0.8180 kcal/mol. The 1,4 van der Waals term of 0.6791 dominates the steric energy. This term is due to the H-H repulsion contribution. Note: The values of the energy terms shown are approximate and can vary slightly based on the type of processor used to calculate them. Dihedral angles To view the value of one of the dihedral angles that contributes to the 1,4 van der Waals contribution: 1. Select the atoms making up the dihedral angle as shown in the figure below by Shift+clicking H(7), C(2), C(1), and H(4) in that order.
57
ChemBioDraw Ultra 12.0
August 2011
2. Go to Structure>Measurements>Display Dihedral Measurement. The displayed angle represents the lowest energy conformation for the ethane model. Using constraint values Entering a value in the Optimal column (in the Measurement table) imposes a constraint on the minimization routine. You are increasing the force constant for the torsional term in the steric energy calculation so that you can optimize to the transition state. 1. Select the Trackball tool. 2. Reorient the model by dragging the X- and Y-axis rotation bars until you have an end-on view. To force a minimization to converge on the transition conformation, set the barrier to rotation: 1. In the Measurement table, type 0 in the Optimal column for the selected dihedral angle and press the Enter key. 2. Go to Calculations>MM2>Minimize Energy. The Minimize Energy dialog box appears. 3. Click Run. When the minimization is complete, the model conforms to the eclipsed structure and the reported energy values appear in the Output window. The energy for this eclipsed conformation is higher relative to the staggered form. The majority of the energy contribution is from the torsional energy and the 1,4 van der Waals interactions. Note: The values of the energy terms shown here are approximate and can vary slightly based on the type of processor used to calculate them. The dihedral angle in the Actual column becomes 0, corresponding to the imposed constraint. The difference in energy between the global minimum (Total, previous calculation) and the transition state (Total, this calculation) is 2.50 kcal/mole, which is in agreement with literature values. To further illustrate points about minimization, delete the value from the Optimal column for the dihedral angle. Then, click the MM2 icon on the Calculation toolbar. After the minimization is complete, you are still at 0 degrees. This is an important consideration for working with the MM2 minimizer. It uses first derivatives of energy to determine the next logical move to lower the energy. However, for saddle points (transition states), the region is fairly flat and the minimizer is satisfied that a minimum has been reached. If you suspect your starting point is not a minimum, try setting the dihedral angle off by about 2 degrees and minimize again.
58
ChemBioDraw Ultra 12.0
August 2011
Example: Cyclohexane In the following example you compare the cyclohexane twist-boat conformation and the chair global minimum. To build a model of cyclohexane: 1. Go to File>New. An empty model window appears. 2. Select the Text Building tool .
3. Click in the model window. A text box appears.
4. Type CH2(CH2)5 and press the Enter key. While there are other, perhaps easier, methods of creating a cyclohexane model, you should use the method described to follow this example. Before minimizing, use the Clean Up Structure command to refine the model. This generally improves the ability of the Minimize Energy command to reach a minimum point. 1. Go to Edit>Select All. 2. Go to Structure>Clean Up. Note: The Clean Up command is very similar to the minimize energy command in that it is a preset, short minimization of the structure. To perform the minimization, go to Calculations>MM2>Minimize Energy and click Run. When the minimization is complete, reorient the model so it appears as in the figure below.
The conformation you converged to is not the well-known chair conformation, which is the global minimum. Instead, the model has converged on a local minimum, the twisted-boat conformation. This is the closest low-energy conformation to your starting conformation. NB. In the above example Show Hydrogen Atoms has been set to Hide.
59
ChemBioDraw Ultra 12.0
August 2011
Had you built this structure using substructures that are already energy minimized, or in the ChemDraw panel, you would be close to the chair conformation. The minimizer does not surmount the saddle point to locate the global minimum, and the closest minimum is sought. The major contributions are from the 1,4 van der Waals and torsional aspects of the model. For cyclohexane, there are six equivalent local minima (twisted-boat), two equivalent global minima (chair), and many transition states (one of which is the boat conformation). Locating the Global Minimum Finding the global minimum is extremely challenging for all but the most simple molecules. It requires a starting conformation which is already in the valley of the global minimum, not in a local minimum valley. The case of cyclohexane is straightforward because you already know that the global minimum is either of the two possible chair conformations. To obtain the new starting conformation, change the dihedrals of the twisted conformation so that they represent the potential energy valley of the chair conformation. The most precise way to alter a dihedral angle is to change its Actual value in the Measurement table when dihedral angles are displayed. An easier way to alter an angle, especially when dealing with a ring, is to move the atoms by dragging, then cleaning up the resulting conformation. To change a dihedral angle: 1. Drag C1 below the plane of the ring. The cursor appears as a box with a hand. 2. Drag C4 above the plane of the ring. During dragging, the bond lengths and angles were deformed. To return them to the optimal values before minimizing, select the model by dragging a box around it with the Select tool, and run Clean Up. Now run the minimization: 1. Go to Calculations>MM2>Minimize Energy and click Run. Allow the minimization to finish. 2. Reorient the model using the rotation bars to see the final chair conformation. Note: The values of the energy terms shown here are approximate and can vary slightly based on the type of processor used to calculate them.
60
ChemBioDraw Ultra 12.0
August 2011
This conformation conformation.
is
about
5.5
kcal/mole
more
stable
than
the
twisted-boat
For molecules more complicated than cyclohexane, where you dont already know what the global minimum is, other methods may be necessary for locating likely starting geometries for minimization. One way of accessing this conformational space of a molecule with large energy barriers is to perform molecular dynamics simulations. This, in effect, heats the molecule, thereby increasing the kinetic energy enough to cross the energetically disfavored transition states.
61
ChemBioDraw Ultra 12.0
August 2011
Docking Models
The Dock command lets you position a fragment in a desired orientation and proximity relative to a second fragment. Each fragment remains rigid during the docking computation. The Dock command is available when two or more distances between atoms in one fragment and atoms in a second fragment are specified. These distances are entered into the Optimal field in the Measurement table. You can use docking to simulate the association of regions of similar lipophilicity and hydrophilicity on two proximate polymer chains. In this tutorial, the Dock command is demonstrated using two polymer chains. Build the first polymer chain 1. 2. 3. 4. Open a new Model window and select the Text Building tool. Click in the model window. A text box appears. Type (AA-mon)3(C2F4)4(AA-mon)3H in the text box. Press the Enter key.
A polyacrylic acid/polytetrafluoroethylene block copolymer appears in the model window. The text, (AA-mon)3, is converted to a polymer segment with three repeat units of acrylic acid. The text, (C2F4)4, is converted to a polymer segment with four repeat units of tetrafluoroethylene. Build a copy of the chain Click in the model window well above and to the right of the first model. When the filled text box appears, press the Enter key. A second polymer molecule appears. Orient the chains 1. Click in the empty space in the model window to deselect any atoms in the model window. 2. Click the down arrow on the Trackball tool to open the rotation dial tool. 3. Select the Y axis, and drag the dial to show 55. Tip: To get exactly 55 you may need to edit the value in the number box. After editing, press the Enter key. The value displayed in the right corner of the dial should be the same as in the number box. Set optimal distances The Optimal distance determines how closely the molecules dock. In this tutorial, you will set the distance to 5. 1. In the Model Explorer (from the View menu), select C(6) in Fragment 1 in the AAmon 2 group. 2. Locate the C(98) atom in Fragment 2 (AAmon 21 group) and CTRL-click to select it. 3. Go to Structure>Measurements>Display Distance Measurement. The Measurement table opens (or becomes active), displaying the C(98)-C(6) pair. 4. Click the Optimal cell. 5. Type 5 and press the Enter key.
62
ChemBioDraw Ultra 12.0
August 2011
The optimal distance between C(6) and C(98) is specified as 5.000. To have a reasonable dock, you must specify at least four atom pairs. Repeat steps 1 through 6 for matching atom pairs throughout the fragments:
Atoms C(1)-C(93) C(98)-C(6) C(104)-C(12) C(108)-C(16) C(22)-C(114) C(28)-C(120) C(34)-C(126) C(133)-C(41) C(45)-C(137) C(50)-C(142)
Actual 21.2034 21.1840 21.2863 21.1957 20.6472 20.7001 20.1410 20.3559 20.3218 20.4350
Optimal 5.0000 5.0000 5.0000 5.0000 5.0000 5.0000 5.0000 5.0000 5.0000 5.0000
Ignore the distances in the Actual cell because they depend on how the second polymer was positioned relative to the first polymer when the second polymer was created. To begin the docking computation: 1. Go to Structure>Dock. The Dock dialog box appears. 2. Type 0.100 for the Minimum RMS Error value and 0.010 for the Minimum RMS Gradient. The docking computation stops when the RMS Error or the RMS Gradient becomes less than the Minimum RMS Error and Minimum RMS Gradient value. 3. Click Display Each Iteration. This allows you to see how much the fragments have moved after each iteration of the docking computation. 4. Click Start. Note that while the docking computation proceeds, one molecule remains stationary and the second molecule moves. To stop the docking computation before it reaches its preset RMS values, click Stop Calculation on the Calculation toolbar. Both docking and recording are stopped. The Status bar displays the values describing each iteration. The relative position of the two fragments or molecules at the start of the docking computation can affect your results. For more accurate results, lower the minimum RMS gradient.
63
ChemBioDraw Ultra 12.0
August 2011
Further Help
ChemBioDraw Help menu
ChemBio3D Help menu
CambridgeSoft Desktop Support http://www.cambridgesoft.com/services/DesktopSupport/KnowledgeBase/FAQ/Default.a spx
64
Вам также может понравиться
- CM 7.2 Manual Session 1Документ141 страницаCM 7.2 Manual Session 1Diana Rey100% (2)
- Practical Process Control for Engineers and TechniciansОт EverandPractical Process Control for Engineers and TechniciansРейтинг: 5 из 5 звезд5/5 (3)
- PIC Microcontroller Projects in C: Basic to AdvancedОт EverandPIC Microcontroller Projects in C: Basic to AdvancedРейтинг: 5 из 5 звезд5/5 (10)
- MATLAB Programming for Biomedical Engineers and ScientistsОт EverandMATLAB Programming for Biomedical Engineers and ScientistsРейтинг: 4.5 из 5 звезд4.5/5 (4)
- Power MillДокумент1 176 страницPower Millyorgf92% (13)
- Lecture Note On Mass SpectrosДокумент22 страницыLecture Note On Mass Spectrosm__rubel96% (27)
- Organic Structural Spectroscopy 2nd Edition Lambert Solutions ManualДокумент7 страницOrganic Structural Spectroscopy 2nd Edition Lambert Solutions Manualkevahung907e100% (28)
- Mass Spectrometry Dry Lab CHM 1021Документ7 страницMass Spectrometry Dry Lab CHM 1021himaОценок пока нет
- ChemDraw TrainingДокумент64 страницыChemDraw Trainingslash_de_law7484Оценок пока нет
- Factorytalk View Machine Edition User'S GuideДокумент587 страницFactorytalk View Machine Edition User'S Guideanon_723873298Оценок пока нет
- Tekla Structure System - TutorialДокумент500 страницTekla Structure System - TutorialJohn Colocon100% (4)
- Thermolib UserManualДокумент334 страницыThermolib UserManualSarmad HussainОценок пока нет
- Viewme Um004 - en e PDFДокумент526 страницViewme Um004 - en e PDFDhavalОценок пока нет
- Argouml User Manual: A Tutorial and Reference DescriptionДокумент394 страницыArgouml User Manual: A Tutorial and Reference DescriptionJosé CarlosОценок пока нет
- Mach4 Screen EditorДокумент33 страницыMach4 Screen EditorluisОценок пока нет
- Microplate Reader RT 2100C MANUALДокумент50 страницMicroplate Reader RT 2100C MANUALHugo Francisco Felipe Caceros100% (1)
- Viewse Um006 - en eДокумент536 страницViewse Um006 - en eMaria Carolina Obregon YaarОценок пока нет
- RICOH MP C2503 User ManualДокумент240 страницRICOH MP C2503 User ManualJahid Masud100% (1)
- BMC Base de Conocimiento LinuxДокумент430 страницBMC Base de Conocimiento LinuxjmandirolaОценок пока нет
- Thermolib GettingStartedДокумент35 страницThermolib GettingStartedArjuna SarumpaetОценок пока нет
- Curso Basico td6Документ135 страницCurso Basico td6Ildefonso Reali CastrejonОценок пока нет
- Intro To Signal Processing 2020Документ494 страницыIntro To Signal Processing 2020QS LОценок пока нет
- Plate-Forme de Modules Modicon x80 - CatalogДокумент71 страницаPlate-Forme de Modules Modicon x80 - CatalogMOHAMED BOUZEBRAОценок пока нет
- Logix Um003 - en PДокумент116 страницLogix Um003 - en Pspe.ot5Оценок пока нет
- 670 Series 2.0 IEC: Operation ManualДокумент112 страниц670 Series 2.0 IEC: Operation ManualLuis YucraОценок пока нет
- 3BSE038018-600 - en System 800xa 6.0 System Guide Functional DescriptionДокумент588 страниц3BSE038018-600 - en System 800xa 6.0 System Guide Functional DescriptionSabzgostar Avande ParsОценок пока нет
- P LLSДокумент56 страницP LLSdeccanchaserОценок пока нет
- Ba Atmocontrol FDA en d30396Документ56 страницBa Atmocontrol FDA en d30396HythamMidaniОценок пока нет
- ArtcamДокумент351 страницаArtcamMilannet Milannet100% (5)
- 3BSE035980-510 - en System 800xa Control 5.1 AC 800M Configuration PDFДокумент694 страницы3BSE035980-510 - en System 800xa Control 5.1 AC 800M Configuration PDFSaada LamОценок пока нет
- Proces rm006 - en P (P - Seq) PDFДокумент72 страницыProces rm006 - en P (P - Seq) PDFMarcioWatanabeОценок пока нет
- Syslib Rm019 en e (P AInDual)Документ52 страницыSyslib Rm019 en e (P AInDual)carbono980Оценок пока нет
- 058-80 BTL-4000 Smart&Premium User Manual EN206Документ72 страницы058-80 BTL-4000 Smart&Premium User Manual EN206Sayed AlmokalyОценок пока нет
- Manual Control Flash - PLUSДокумент75 страницManual Control Flash - PLUSglzmtzjonathanОценок пока нет
- CBECC Com UserManualДокумент229 страницCBECC Com UserManualbradalbiОценок пока нет
- 1794-FLEX IO High-Density Analog ModulesДокумент90 страниц1794-FLEX IO High-Density Analog ModulesBenavides NestorОценок пока нет
- FeatureCAM Whats New 2012 R3Документ153 страницыFeatureCAM Whats New 2012 R3anasrl2006Оценок пока нет
- 3BSE042835R5001 CIO LibObjDesignДокумент90 страниц3BSE042835R5001 CIO LibObjDesignCristi CrseОценок пока нет
- Viewse Um006 - en e (001 112)Документ112 страницViewse Um006 - en e (001 112)NeuОценок пока нет
- Arena Template Developer's GuideДокумент286 страницArena Template Developer's Guidejhonny_otiОценок пока нет
- Controlador de PLCДокумент378 страницControlador de PLCZerpa Rodriguez YancarlosОценок пока нет
- Manual de Program. CPU Mitsubishi - R04CPUДокумент156 страницManual de Program. CPU Mitsubishi - R04CPUJefferson MisaelОценок пока нет
- Atg Service Center Ui Prog GuideДокумент158 страницAtg Service Center Ui Prog GuideArunОценок пока нет
- SALOME TutorialДокумент82 страницыSALOME TutorialPouya Ghasemi0% (1)
- Pipesim User GuideДокумент1 014 страницPipesim User GuideToufikNechОценок пока нет
- Ftac gr002 - en eДокумент110 страницFtac gr002 - en eCarlos Andres Morales PovedaОценок пока нет
- Niisim, A Simulator For Computer Engineering Education: Emil BäckströmДокумент66 страницNiisim, A Simulator For Computer Engineering Education: Emil BäckströmtptuyenОценок пока нет
- ChemWell R6Документ226 страницChemWell R6Claudio BiolcatiОценок пока нет
- Template Developer's Guide PDFДокумент295 страницTemplate Developer's Guide PDFMariangeles XitumulОценок пока нет
- UTG 38 DL +user ManualДокумент318 страницUTG 38 DL +user ManualShahbaz KhanОценок пока нет
- AutomotiveCockpitDesign2020 PDFДокумент85 страницAutomotiveCockpitDesign2020 PDFpetrishiaОценок пока нет
- Introduction of an Employee-Led Training System at Stage Entertainment Germany: A Qualitative StudyОт EverandIntroduction of an Employee-Led Training System at Stage Entertainment Germany: A Qualitative StudyОценок пока нет
- Electronics from the Ground Up: Learn by Hacking, Designing, and InventingОт EverandElectronics from the Ground Up: Learn by Hacking, Designing, and InventingРейтинг: 3.5 из 5 звезд3.5/5 (2)
- Web 2.0 and the Health Care Market: Health Care in the era of Social Media and the modern InternetОт EverandWeb 2.0 and the Health Care Market: Health Care in the era of Social Media and the modern InternetОценок пока нет
- Ascen Zi 2013Документ9 страницAscen Zi 2013Brahma Hakim Yuanda HutabaratОценок пока нет
- N. Sabila, P. M.Mwangi, P. Kareru and G. Thiong'o Department Chemistry, Jomo Kenyatta University of Agriculture and Technology, Nairobi, KenyaДокумент8 страницN. Sabila, P. M.Mwangi, P. Kareru and G. Thiong'o Department Chemistry, Jomo Kenyatta University of Agriculture and Technology, Nairobi, KenyaAli DandamunОценок пока нет
- Intermediate Organic ChemistryДокумент428 страницIntermediate Organic ChemistryMichael S. Leonard100% (3)
- Mass Spectrometric Studies On Selective Androgen Receptor Modulators (Sarms) Using Electron Ionization and Electrospray Ionization/Collision-Induced DissociationДокумент12 страницMass Spectrometric Studies On Selective Androgen Receptor Modulators (Sarms) Using Electron Ionization and Electrospray Ionization/Collision-Induced DissociationA O 10Оценок пока нет
- Reactivity of Oximes For Diverse Methodologies and Synthetic ApplicationsДокумент13 страницReactivity of Oximes For Diverse Methodologies and Synthetic ApplicationsAnahí TessaОценок пока нет
- M.SC Chemistry Curriculum Syllabus 2018 19Документ68 страницM.SC Chemistry Curriculum Syllabus 2018 19krishna kumarОценок пока нет
- Mass Spectroscopy - PPTX Paper 2Документ27 страницMass Spectroscopy - PPTX Paper 2Prerana RaneОценок пока нет
- Stanford AIДокумент6 страницStanford AInguyenhaquanОценок пока нет
- Mass Spectrometry PDFДокумент4 страницыMass Spectrometry PDFFran LeeОценок пока нет
- Fragmentations PaternДокумент22 страницыFragmentations PaternkhadijahhannahОценок пока нет
- Mass Spec LectureДокумент41 страницаMass Spec LectureAsim Alaa Al SalehiОценок пока нет
- Fragmentation in Mass Spectrometry: - Introduction - Ionization - SeparationДокумент38 страницFragmentation in Mass Spectrometry: - Introduction - Ionization - SeparationRøccø RäjОценок пока нет
- Test Bank For Organic Chemistry 8th Edition McmurryДокумент18 страницTest Bank For Organic Chemistry 8th Edition McmurryAnthonyRogersydtfp100% (65)
- J. Agric. Food Chem. 2005, 53, 9010-9016Документ8 страницJ. Agric. Food Chem. 2005, 53, 9010-9016Jatyr OliveiraОценок пока нет
- Mass Spectroscopy.Документ23 страницыMass Spectroscopy.Ashley BedassieОценок пока нет
- Ch. 6 Mass SpectometryДокумент45 страницCh. 6 Mass Spectometryrazan ghuweiriОценок пока нет
- 5.1 Mass Spectrometry FactfileДокумент5 страниц5.1 Mass Spectrometry FactfileFasiha RazaОценок пока нет
- Mass Spectroscopy: - : Presented By:-Amruta S. Sambarekar 1 Year M.Pharm Dept. of Pharmaceutics M M C P, BelgaumДокумент56 страницMass Spectroscopy: - : Presented By:-Amruta S. Sambarekar 1 Year M.Pharm Dept. of Pharmaceutics M M C P, BelgaumSudeep_Kolhar_5702Оценок пока нет
- Molecular Mass Spectroscopy:: HistoryДокумент43 страницыMolecular Mass Spectroscopy:: Historyramesh pokhrelОценок пока нет
- T SC 2551110 A Level Chemistry Mass Spectrometry Study Guide - Ver - 3Документ7 страницT SC 2551110 A Level Chemistry Mass Spectrometry Study Guide - Ver - 3sara.u1925Оценок пока нет
- Ait Raman Ms NMRДокумент56 страницAit Raman Ms NMRAditya YadavОценок пока нет
- Mass SpectrosДокумент36 страницMass SpectrosErsya NurriaОценок пока нет
- SEM-III - Organic Spectroscopy and Rearrangements - Unit-III (Mass Spectroscopy)Документ47 страницSEM-III - Organic Spectroscopy and Rearrangements - Unit-III (Mass Spectroscopy)PG ChemistryОценок пока нет
- International Journal of Mass Spectrometry - 2019Документ6 страницInternational Journal of Mass Spectrometry - 2019luiz eduardoОценок пока нет
- An04 Massenspektroskopie Skript VolmerДокумент160 страницAn04 Massenspektroskopie Skript VolmerFasiha RazaОценок пока нет
- STEM Success Center: CHEM 201 WorksheetДокумент3 страницыSTEM Success Center: CHEM 201 WorksheetFasiha RazaОценок пока нет
- New Developments in The Chemistry of Metal-Acetylacetonate ComplexesДокумент37 страницNew Developments in The Chemistry of Metal-Acetylacetonate ComplexesDzul HadisОценок пока нет