Академический Документы
Профессиональный Документы
Культура Документы
Lab Manual 3
Загружено:
syazaismailАвторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Lab Manual 3
Загружено:
syazaismailАвторское право:
Доступные форматы
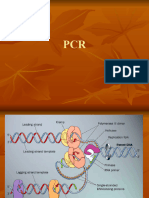
Lab 3: Polymerase Chain Reaction (PCR) of Target DNA Fragment Introduction
In molecular biology, the polymerase chain reaction (PCR) is a technique to amplify a single or few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence. The method relies on thermal cycling, consisting of cycles of repeated heating and cooling reactions for DNA melting, annealing and enzymatic replication of the DNA. Primers (short DNA fragments) containing sequences complementary to the target region along with a thermostable DNA polymerase are key components to enable selective and repeated amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. The PCR usually consists of a series of 20 to 40 repeated temperature changes called cycles; each cycle typically consists of 2-3 discrete temperature steps. Most commonly PCR is carried out with cycles that have three temperature steps:
Denaturation step: This step is the first regular cycling event and consists of heating the reaction to 9498 C for 2030 seconds. It causes DNA melting of the DNA template by disrupting the hydrogen bonds between complementary bases, yielding single strands of DNA. Annealing step: The reaction temperature is lowered to 5065 C for 2040 seconds allowing annealing of the primers to the single-stranded DNA template. Typically the Ta (annealing temperature) is about 3-5 degrees Celsius below the Tm (melting temperature) of the primers used. Extension/elongation step: The temperature at this step depends on the DNA polymerase used; Taq DNA polymerase has its optimum activity temperature at 75 80 C, and commonly a temperature of 72 C is used with this enzyme. At this step the DNA polymerase synthesizes a new DNA strand complementary to the DNA template strand by adding dNTPs that are complementary to the template in 5' to 3' direction.
In PCR, a pair of primer is used to determine the DNA fragment to be amplified by the PCR process. The length of primers is usually not more than 30 nucleotides, and they match exactly the beginning and the end of the DNA fragment to be amplified. Pairs of primers should have similar Tm, as annealing in a PCR occurs for both simultaneously. The following formula is used to calculate the Tm of DNA primer: Tm = 62.3oC + 0.41oC (% G+C) 500/length
Methodology
1. Label 0.5 ml microtube according to your group number. 2. Prepare the PCR mixture as indicated below (transfer the reagent according to the sequence, a to h): Reagent a. Distilled water b. 10X buffer c. MgCl2 (25 mM) d. dNTPs (2.5 mM) e. Forwardprimer (10 M) f. Reverse primer (10 M) g. Plasmid h.Taq DNA polymerase Total volume: Volume (l) 18 2.5 1.5 0.5 0.5 0.5 1 0.5 25 Final concentration ? ? ? ? ? -
3. Calculate the final concentration of each reagent.
4. Conduct the PCR as follows: Reaction Initial denaturation Denature Annealing Extension Final extension Temperature (oC) 94 94 55 72 72 Time 3 min 30 sec 50 sec 1 min 30 sec 7 min
5. Run the amplified DNA fragment on an agarose gel electrophoresis with an appropriate DNA marker.
Questions
1. In the conduct of PCR experiment two extra reactions, namely initial denaturation and final extension are included? Give reasons why these two reactions are used.
2. If a student use the following DNA primers in PCR. Calculate the Tm of both primers. Forward primer 5' gcc agg gcc ctt att atg tct tcc gta ttc g 3' Reverse primer 5' ata tca ata ccc cca gtc gga ggt gtc g 3'
3. Based on the X gene sequence given to you, design a forward and reverse primers that amplified the whole sequence. Calculate the Tm of both primers and suggest the appropriate Tm to be used in PCR.
Appendix I Vector Map, DNA Sequence and Primers
Forward primer,
5 GCTGATGTCGACGAGCGTAC 3
The shaded area is the sequence for Sal I endonuclease recognition site
Reverse primer = 5 GTTATCGCTTAGCCGACGCC 3
The shaded area is the sequence for Blp I endonuclease recognition site (Note: remember, this the reverse primer, intended to copy and amplify the complementary strand)
The entire DNA sequence for a X gene in pBAD vector. (The first codon is in green and underlined whereas the last codon is in red and also underlined)
AAGAAACCAATTGTCCATATTGCATCAGACATTGCCGTCACTGCGTCTTTTACTGGCTCTTCTCGCTAACC AAACCGGTAACCCCGCTTATTAAAAGCATTCTGTAACAAAGCGGGACCAAAGCCATGACAAAAACGCGTAA CAAAAGTGTCTATAATCACGGCAGAAAAGTCCACATTGATTATTTGCACGGCGTCACACTTTGCTATGCCA TAGCATTTTTATCCATAAGATTAGCGGATCCTACCTGACGCTTTTTATCGCAACTCTCTACTGTTTCTCCA TACCCGTTTTTTTGGGCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATACCCATGGGCTCTG GATCCGGTGATGACGATGACAAGCTCGCCCTTATGAAAGCGATCTTAATCCCATTTTTATCTCTTCTGATT CCGTTAACCCCGCAATCTGCATTCGCTCAGAGTGAGCCGGAGCTGAAGCTGGAAAGTGTGGTGATTGTCAG TCGTCATGGTGTGCGTGCTCCAACCAAGGCCACGCAACTGATGCAGGATGTCACCCCAGACGCATGGCCAA CCTGGCCGGTAAAACTGGGTTGGCTGACACCGCGCGGTGGTGAGCTAATCGCCTATCTCGGACATTACCAA CGCCAGCGTCTGGTAGCCGACGGATTGCTGGCGAAAAAGGGCTGCCCGCAGTCTGGTCAGGTCGCGATTAT TGCTGATGTCGACGAGCGTACCCGTAAAACAGGCGAAGCCTTCGCCGCCGGGCTGGCACCTGACTGTGCAA TAACCGTACATACCCAGGCAGATACGTCCAGTCCCGATCCGTTATTTAATCCTCTAAAAACTGGCGTTTGC CAACTGGATAACGCGAACGTGACTGACGCGATCCTCAGCAGGGCAGGAGGGTCAATTGCTGACTTTACCGG GCATCGGCAAACGGCGTTTCGCGAACTGGAACGGGTGCTTAATTTTCCGCAATCAAACTTGTGCCTTAAAC GTGAGAAACAGGACGAAAGCTGTTCATTAACGCAGGCATTACCATCGGAACTCAAGGTGAGCGCCGACAAT GTCTCATTAACCGGTGCGGTAAGCCTCGCATCAATGCTGACGGAGATATTTCTCCTGCAACAAGCACAGGG AATGCCGGAGCCGGGGTGGGGAAGGATCACCGATTCACACCAGTGGAACACCTTGCTAAGTTTGCATAACG CGCAATTTTATTTGCTACAACGCACGCCAGAGGTTGCCCGCAGCCGCGCCACCCCGTTATTAGATTTGATC AAGACAGCGTTGACGCCCCATCCACCGCAAAAACAGGCGTATGGTGTGACATTACCCACTTCAGTGCTGTT TATCGCCGGACACGATACTAATCTGGCAAATCTCGGCGGCGCACTGGAGCTCAACTGGACGCTTCCCGGTC AGCCGGATAACACGCCGCCAGGTGGTGAACTGGTGTTTGAACGCTGGCGTCGGCTAAGCGATAACAGCCAG TGGATTCAGGTTTCGCTGGTCTTCCAGACTTTACAGCAGATGCGTGATAAAACGCCGCTGTCATTAAATAC GCCGCCCGGAGAGGTGAAACTGACCCTGGCAGGATGTGAAGAGCGAAATGCGCAGGGCATGTGTTCGTTGG CAGGTTTTACGCAAATCGTGAATGAAGCACGCATACCGGCGTGCAGTTTGTAAAAGGGCGAGCTTGAAGGT AAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGCGTACCGGTCATCATCACCATCACCATTGAGT TTAAACGGTCTCCAGCTTGGCTGTTTTGGCGGATGAGAGAAGATTTTCAGCCTGATACAGATTAAATCAGA ACGCAGAAGCGGTCTGATAAAACAGAATTTGCCTGGCGGCAGTAGCGCGGTGGTCCCACCTGACCCCATGC CGAACTCAGAAGTGAAACGCCGTAGCGCCGATGGTAGTGTGGGGTCTCCCCATGCGAGAGTAGGGAACTGC
CAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGA ACGCTCTCCTGAGTAGGACAAATCCGCCGGGAGCGGATTTGAACGTTGCGAAGCAACGGCCCGGAGGGTGG CGGGCAGGACGCCCGCCATAAACTGCCAGGCATCAAATTAAGCAGAAGGCCATCCTGACGGATGGCCTTTT TGCGTTTCTACAAACTCTTTTTGTTTATTTTTCTAAATACATTCAAATATGTATCCGCTCATGAGACAATA ACCCTGATAAATGCTTCAATAATATTGAAAAAGGAAGAGTATGAGTATTCAACATTTCCGTGTCGCCCTTA TTCCCTTTTTTGCGGCATTTTGCCTTCCTGTTTTTGCTCACCCAGAAACGCTGGTGAAAGTAAAAGATGCT GAAGATCAGTTGGGTGCACGAGTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGTTT TCGCCCCGAAGAACGTTTTCCAATGATGAGCACTTTTAAAGTTCTGCTATGTGGCGCGGTATTATCCCGTG TTGACGCCGGGCAAGAGCAACTCGGTCGCCGCATACACTATTCTCAGAATGACTTGGTTGAGTACTCACCA GTCACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTATGCAGTGCTGCCATAACCATGAGTGA TAACACTGCGGCCAACTTACTTCTGACAACGATCGGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAACA TGGGGGATCATGTAACTCGCCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGT GACACCACGATGCCTGTAGCAATGGCAACAACGTTGCGCAAACTATTAACTGGCGAACTACTTACTCTAGC TTCCCGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCGGCCCTTC CGGCTGGCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGCGGTATCATTGCAGCACTG GGGCCAGATGGTAAGCCCTCCCGTATCGTAGTTATCTACACGACGGGGAGTCAGGCAACTATGGATGAACG AAATAGACAGATCGCTGAGATAGGTGCCTCACTGATTAAGCATTGGTAACTGTCAGACCAAGTTTACTCAT ATATACTTTAGATTGATTTAAAACTTCATTTTTAATTTAAAAGGATCTAGGTGAAGATCCTTTTTGATAAT CTCATGACCAAAATCCCTTAACGTGAGTTTTCGTTCCACTGAGCGTCAGACCCCGTAGAAAAGATCAAAGG ATCTTCTTGAGATCCTTTTTTTCTGCGCGTAATCTGCTGCTTGCAAACAAAAAAACCACCGCTACCAGCGG TGGTTTGTTTGCCGGATCAAGAGCTACCAACTCTTTTTCCGAAGGTAACTGGCTTCAGCAGAGCGCAGATA CCAAATACTGTCCTTCTAGTGTAGCCGTAGTTAGGCCACCACTTCAAGAACTCTGTAGCACCGCCTACATA CCTCGCTCTGCTAATCCTGTTACCAGTGGCTGCTGCCAGTGGCGATAAGTCGTGTCTTACCGGGTTGGACT CAAGACGATAGTTACCGGATAAGGCGCAGCGGTCGGGCTGAACGGGGGGTTCGTGCACACAGCCCAGCTTG GAGCGAACGACCTACACCGAACTGAGATACCTACAGCGTGAGCTATGAGAAAGCGCCACGCTTCCCGAAGG GAGAAAGGCGGACAGGTATCCGGTAAGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGG GAAACGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGC TCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTG GCCTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGT GAGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAAGAGCGC CTGATGCGGTATTTTCTCCTTACGCATCTGTGCGGTATTTCACACCGCATATGGTGCACTCTCAGTACAAT CTGCTCTGATGCCGCATAGTTAAGCCAGTATACACTCCGCTATCGCTACGTGACTGGGTCATGGCTGCGCC CCGACACCCGCCAACACCCGCTGACGCGCCCTGACGGGCTTGTCTGCTCCCGGCATCCGCTTACAGACAAG CTGTGACCGTCTCCGGGAGCTGCATGTGTCAGAGGTTTTCACCGTCATCACCGAAACGCGCGAGGCAGCAG ATCAATTCGCGCGCGAAGGCGAAGCGGCATGCATAATGTGCCTGTCAAATGGACGAAGCAGGGATTCTGCA AACCCTATGCTACTCCGTCAAGCCGTCAATTGTCTGATTCGTTACCAATTATGACAACTTGACGGCTACAT CATTCACTTTTTCTTCACAACCGGCACGGAACTCGCTCGGGCTGGCCCCGGTGCATTTTTTAAATACCCGC GAGAAATAGAGTTGATCGTCAAAACCAACATTGCGACCGACGGTGGCGATAGGCATCCGGGTGGTGCTCAA AAGCAGCTTCGCCTGGCTGATACGTTGGTCCTCGCGCCAGCTTAAGACGCTAATCCCTAACTGCTGGCGGA AAAGATGTGACAGACGCGACGGCGACAAGCAAACATGCTGTGCGACGCTGGCGATATCAAAATTGCTGTCT GCCAGGTGATCGCTGATGTACTGACAAGCCTCGCGTACCCGATTATCCATCGGTGGATGGAGCGACTCGTT AATCGCTTCCATGCGCCGCAGTAACAATTGCTCAAGCAGATTTATCGCCAGCAGCTCCGAATAGCGCCCTT CCCCTTGCCCGGCGTTAATGATTTGCCCAAACAGGTCGCTGAAATGCGGCTGGTGCGCTTCATCCGGGCGA AAGAACCCCGTATTGGCAAATATTGACGGCCAGTTAAGCCATTCATGCCAGTAGGCGCGCGGACGAAAGTA AACCCACTGGTGATACCATTCGCGAGCCTCCGGATGACGACCGTAGTGATGAATCTCTCCTGGCGGGAACA GCAAAATATCACCCGGTCGGCAAACAAATTCTCGTCCCTGATTTTTCACCACCCCCTGACCGCGAATGGTG AGATTGAGAATATAACCTTTCATTCCCAGCGGTCGGTCGATAAAAAAATCGAGATAACCGTTGGCCTCAAT CGGCGTTAAACCCGCCACCAGATGGGCATTAAACGAGTATCCCGGCAGCAGGGGATCATTTTGCGCTTCAG CCATACTTTTCATACTCCCGCCATTCAGAG
Fig 1: pBAD=TOPO map (adapted from pBAD-TOPO Expression Kit manual, INVITROGEN)-
Вам также может понравиться
- PCRДокумент34 страницыPCRSumitОценок пока нет
- Polyme Rase Chain Reactio N: Course Code: 501Документ23 страницыPolyme Rase Chain Reactio N: Course Code: 501Humayun ArshadОценок пока нет
- PCRДокумент42 страницыPCRNopiyana PujiastutiОценок пока нет
- Analiza PCR - ThermocyclerДокумент7 страницAnaliza PCR - Thermocyclerionescu_ion_3Оценок пока нет
- PCR and Agarose Gel ElectrophoresisДокумент6 страницPCR and Agarose Gel ElectrophoresisÍrisОценок пока нет
- Molecular Diagnostics Techniques Polymerase Chain ReactionДокумент7 страницMolecular Diagnostics Techniques Polymerase Chain ReactionJamesОценок пока нет
- MKBS221. PRAC 5 REPORT Last OneДокумент11 страницMKBS221. PRAC 5 REPORT Last OneNOMKHULEKO ALICEОценок пока нет
- Principle of The PCR: This Technique Is Invented in 1984 by Kary Mulis. The Purpose of A PCRДокумент11 страницPrinciple of The PCR: This Technique Is Invented in 1984 by Kary Mulis. The Purpose of A PCRantesar reheemОценок пока нет
- Polymerase Chain ReactionДокумент5 страницPolymerase Chain ReactionAaryan DeswalОценок пока нет
- Radio Immuno As SayДокумент6 страницRadio Immuno As SayhardmanpersonОценок пока нет
- Polymerase Chain Reaction (PCR)Документ21 страницаPolymerase Chain Reaction (PCR)greateinsteinОценок пока нет
- PCR ManualДокумент6 страницPCR ManualjiaОценок пока нет
- Polymerase Chain Reaction (PCR)Документ37 страницPolymerase Chain Reaction (PCR)Haleema SultanОценок пока нет
- PCR Lecture HURO Zador Final SmallДокумент59 страницPCR Lecture HURO Zador Final SmallvsripathirajaОценок пока нет
- Polymerase Chain ReactionДокумент13 страницPolymerase Chain Reactionvamshibommi3477Оценок пока нет
- PCR Overview GoldBioДокумент1 страницаPCR Overview GoldBioKIARAH EUNIZE GICAОценок пока нет
- Polymerase Chain ReactionДокумент47 страницPolymerase Chain ReactionshvnagaОценок пока нет
- PCR LectureДокумент18 страницPCR LecturePagla HowaОценок пока нет
- Polymerase Chain ReactionДокумент16 страницPolymerase Chain ReactionIshikaОценок пока нет
- Polymerase Chain Reaction: Salwa Hassan TeamaДокумент56 страницPolymerase Chain Reaction: Salwa Hassan TeamaSamiksha SharmaОценок пока нет
- DNA Amplification 2020Документ5 страницDNA Amplification 2020Blameless ArikoОценок пока нет
- Polymerase Chain ReactionДокумент27 страницPolymerase Chain ReactionundalliОценок пока нет
- TYPES OF PCR - Dr. De-1 PDFДокумент33 страницыTYPES OF PCR - Dr. De-1 PDFJAICHANDRUОценок пока нет
- Bio Project-Polymerase Chain ReactionДокумент21 страницаBio Project-Polymerase Chain ReactionS.AbiniveshОценок пока нет
- Components of PCRДокумент3 страницыComponents of PCRpmc9q544vwОценок пока нет
- Ex No.-03 Polymerase Chain ReactionДокумент4 страницыEx No.-03 Polymerase Chain ReactionSujit ShandilyaОценок пока нет
- The Polymerase Chain Reaction and Sequence-Tagged SitesДокумент7 страницThe Polymerase Chain Reaction and Sequence-Tagged SitesPisi MicaОценок пока нет
- Pta, Which Encodes The Enzyme Phosphoacetyltransferase, Revealed The Presence of An Mboi RestrictionДокумент5 страницPta, Which Encodes The Enzyme Phosphoacetyltransferase, Revealed The Presence of An Mboi RestrictionANNAALTSHULERОценок пока нет
- The Olymerase Hain Eaction: Zhang-Haifeng Department of BiochemistryДокумент42 страницыThe Olymerase Hain Eaction: Zhang-Haifeng Department of Biochemistryapi-19916399Оценок пока нет
- BB2111 PCR TOBEPRINTEDДокумент7 страницBB2111 PCR TOBEPRINTEDapi-3839553Оценок пока нет
- Polymerase Chain ReactionДокумент19 страницPolymerase Chain ReactionKamila Haidari100% (1)
- Polymerase Chain Reaction - WikipediaДокумент36 страницPolymerase Chain Reaction - Wikipedia93428ZEFHDОценок пока нет
- The Polymerase Chain Reaction (PCR)Документ50 страницThe Polymerase Chain Reaction (PCR)Deana NamirembeОценок пока нет
- PCR An OverviewДокумент41 страницаPCR An OverviewnassradabourОценок пока нет
- PCRДокумент5 страницPCROh RoopzОценок пока нет
- PCRДокумент35 страницPCRk5zo7jОценок пока нет
- Nucleic Acid Manipulation: The Polymerase Chain ReactionДокумент22 страницыNucleic Acid Manipulation: The Polymerase Chain Reactionamor_panopioОценок пока нет
- Polymerase Chain Reaction ProtocolДокумент14 страницPolymerase Chain Reaction ProtocolDespoina ChatziОценок пока нет
- Module 4 - PCRДокумент12 страницModule 4 - PCRnavneetОценок пока нет
- Tutorial 6 Molecular TechniquesДокумент4 страницыTutorial 6 Molecular TechniquesJason LeeОценок пока нет
- PCR Labwork 2 ENGДокумент4 страницыPCR Labwork 2 ENGmigas1996Оценок пока нет
- Polymerase Chain ReactionДокумент14 страницPolymerase Chain ReactionDavidMugambi100% (1)
- Polymerase Chain Reaction (PCR) : Evros Polydorou April 2018Документ11 страницPolymerase Chain Reaction (PCR) : Evros Polydorou April 2018Evros PolidorouОценок пока нет
- PCRДокумент3 страницыPCRarian.boostani2002Оценок пока нет
- Lab 21A and 21BДокумент8 страницLab 21A and 21BLateesha ThomasОценок пока нет
- Polymerase Chain ReactionДокумент34 страницыPolymerase Chain Reactionmokshgoyal2597100% (4)
- Polymerase Chain Reaction: Presented By: Saurabh Maolanker RD University Jabalpur (M.P.)Документ14 страницPolymerase Chain Reaction: Presented By: Saurabh Maolanker RD University Jabalpur (M.P.)Saurabh MavlankarОценок пока нет
- PCR1Документ4 страницыPCR1Hina NoorОценок пока нет
- Aula TBM5 2022-2023 - EnglishДокумент29 страницAula TBM5 2022-2023 - EnglishDiogo Botelho Medeiros BrilhanteОценок пока нет
- Polymerase Chain ReactionДокумент8 страницPolymerase Chain ReactionAdigun TaofeeqatОценок пока нет
- Topic:: Polymerase Chain Reaction (PCR) Memona Aslam Manisha Mahboob Husnain Javed Iqra Praveen Mam Tehreem 3rd BiologyДокумент23 страницыTopic:: Polymerase Chain Reaction (PCR) Memona Aslam Manisha Mahboob Husnain Javed Iqra Praveen Mam Tehreem 3rd BiologyTehreem FatimaОценок пока нет
- PCR RTPCR, QPCRДокумент88 страницPCR RTPCR, QPCRshravani sahuОценок пока нет
- Polymerase Chain ReactionДокумент13 страницPolymerase Chain Reactionahmad ghОценок пока нет
- Polymerase Chain Reaction: Mario Walean, M.SiДокумент29 страницPolymerase Chain Reaction: Mario Walean, M.SiMutiara SuprihantoОценок пока нет
- Submitted By: Akshat Goel Xii - C R.NoДокумент17 страницSubmitted By: Akshat Goel Xii - C R.NoAkshat GoelОценок пока нет
- What Is PCRДокумент4 страницыWhat Is PCRPARRU xYTОценок пока нет
- Polymerase Chain Reaction (PCR) - Principle, Steps, ApplicationsДокумент5 страницPolymerase Chain Reaction (PCR) - Principle, Steps, ApplicationsMokhtarCheikhОценок пока нет
- Polymerase Chain ReactionДокумент12 страницPolymerase Chain ReactionSaswat MohapatraОценок пока нет
- Gel Electrophoresis of ProteinsОт EverandGel Electrophoresis of ProteinsMichael J DunnОценок пока нет
- Temperature-Responsive Polymers: Chemistry, Properties, and ApplicationsОт EverandTemperature-Responsive Polymers: Chemistry, Properties, and ApplicationsОценок пока нет
- CapsulesДокумент6 страницCapsulessyazaismailОценок пока нет
- Introduction Fermentech 1Документ3 страницыIntroduction Fermentech 1syazaismailОценок пока нет
- Procedures Bioproduct Lab 1Документ2 страницыProcedures Bioproduct Lab 1syazaismailОценок пока нет
- ConclusionДокумент1 страницаConclusionsyazaismailОценок пока нет
- Intro BioseparationДокумент2 страницыIntro BioseparationsyazaismailОценок пока нет
- RESULT Discussion Spray DryerДокумент7 страницRESULT Discussion Spray Dryersyazaismail100% (1)
- LAB Manual 2Документ4 страницыLAB Manual 2syazaismailОценок пока нет
- Introduction To Capsule FillingДокумент3 страницыIntroduction To Capsule FillingsyazaismailОценок пока нет
- Acid Base TitrationДокумент16 страницAcid Base TitrationsyazaismailОценок пока нет
- LAB Manual 1Документ2 страницыLAB Manual 1syazaismailОценок пока нет
- Assignment 1Документ1 страницаAssignment 1syazaismailОценок пока нет
- Lecture5 - Gene Functions - Proteins and EnzymesДокумент25 страницLecture5 - Gene Functions - Proteins and EnzymesGideon CavidaОценок пока нет
- PCRДокумент5 страницPCROh RoopzОценок пока нет
- Transcription and ModificationДокумент23 страницыTranscription and ModificationWarlin FernandezОценок пока нет
- Lecture 3-Enzyme For DNA Manipulation in Recombinant DNAДокумент42 страницыLecture 3-Enzyme For DNA Manipulation in Recombinant DNANurfarahain ZolkeflyОценок пока нет
- Cell 1981 Cech PDFДокумент10 страницCell 1981 Cech PDFSpellkingОценок пока нет
- 5 - BIO-Identify The Role of DNA and RNA in Protein SynthesisДокумент3 страницы5 - BIO-Identify The Role of DNA and RNA in Protein SynthesisCarlo ThornappleОценок пока нет
- Molecular Basis of Inheritance, Class 12, Cbse NotesДокумент33 страницыMolecular Basis of Inheritance, Class 12, Cbse NotesSubho Bhattacharya50% (4)
- Avatics 2017 03-02-15!34!57protein Synthesis Worksheet PracticeДокумент2 страницыAvatics 2017 03-02-15!34!57protein Synthesis Worksheet PracticeMiguel BernalОценок пока нет
- Three Day Gel LabДокумент8 страницThree Day Gel Labianzhou1Оценок пока нет
- Genes Were Known To : 1. Carry Information From One Generation To The NextДокумент30 страницGenes Were Known To : 1. Carry Information From One Generation To The NextGissele AbolucionОценок пока нет
- 1 6-+DNA+Powerpoint+and+Study+Guide PDFДокумент29 страниц1 6-+DNA+Powerpoint+and+Study+Guide PDFARCHANA DASARIОценок пока нет
- Cut and PasteДокумент2 страницыCut and PasteKelsey PerreaultОценок пока нет
- Next-Generation Sequencing Technologies: An Overview: Taishan Hu, Nilesh Chitnis, Dimitri Monos, Anh DinhДокумент11 страницNext-Generation Sequencing Technologies: An Overview: Taishan Hu, Nilesh Chitnis, Dimitri Monos, Anh DinhNina FajsОценок пока нет
- 4th Week 4.2 LAS Science 10Документ8 страниц4th Week 4.2 LAS Science 10James YamОценок пока нет
- Lesson 1.7 Learning Task WorksheetДокумент6 страницLesson 1.7 Learning Task WorksheetRayan abdallaОценок пока нет
- ٣٢٥ Molecular Biology Sabah Linjawi ١Документ15 страниц٣٢٥ Molecular Biology Sabah Linjawi ١Zainab RaikОценок пока нет
- DNA ReplicationДокумент2 страницыDNA ReplicationFrancoise AngeliqueОценок пока нет
- 12 3 PWPT PDFДокумент22 страницы12 3 PWPT PDFapi-262378640Оценок пока нет
- Chapter 9 - DNA Structure and OrganizationДокумент33 страницыChapter 9 - DNA Structure and OrganizationIka BakarОценок пока нет
- Dna FingerprintingДокумент3 страницыDna FingerprintingkriedОценок пока нет
- Principle, Analysis, Application and Challenges of Next-Generation Sequencing: A ReviewДокумент30 страницPrinciple, Analysis, Application and Challenges of Next-Generation Sequencing: A ReviewShahid HanifОценок пока нет
- Nucleic Acids ExamДокумент3 страницыNucleic Acids ExamAlbert TayagОценок пока нет
- Eukaryotic Genome Complexity - Learn Science at ScitableДокумент4 страницыEukaryotic Genome Complexity - Learn Science at ScitableAnder ManaresОценок пока нет
- Polymerase Chain ReactionДокумент5 страницPolymerase Chain ReactionAman MishraОценок пока нет
- Revised TERM PAPERДокумент19 страницRevised TERM PAPERVikal RajputОценок пока нет
- Fundamentals of Biochemistry Life at The Molecular Level 5th Edition Voet Test BankДокумент28 страницFundamentals of Biochemistry Life at The Molecular Level 5th Edition Voet Test BankGeorgeCobbjgbcs100% (15)
- Biopolymers LectureДокумент23 страницыBiopolymers LectureNneka Okafor-EzeaniОценок пока нет
- Mmoi BC SbaДокумент15 страницMmoi BC SbaYoon YoonОценок пока нет
- Biology Final ExamДокумент7 страницBiology Final ExamJillian FajardoОценок пока нет
- Science: Quarter 3 Self-Learning Module 3 Central Dogma of The Transfer of Genetic InformationДокумент16 страницScience: Quarter 3 Self-Learning Module 3 Central Dogma of The Transfer of Genetic InformationKaye ViolaОценок пока нет