Академический Документы
Профессиональный Документы
Культура Документы
Dna Notes (3&7) - Complete
Загружено:
cgao30Авторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Dna Notes (3&7) - Complete
Загружено:
cgao30Авторское право:
Доступные форматы
IB Biology 2013
1
DNA Notes
3.3 DNA structure
3.3.1 Outline DNA nucleotide structure in terms of sugar (deoxyribose), base and phosphate (2)
A DNA nucleotide has a deoxyribose (pentose) sugar covalently bonded to a nitrous base and
phosphate group
3.3.2 State the names of the four bases in DNA (1)
The four bases in DNA are:
1. Adenine
2. Cytosine
3. Guanine
4. Thymine
3.3.3 Outline how DNA nucleotides are linked together by covalent bonds into a single strand (2)
The DNA nucleotides are linked together by covalent bonds (phosphodiester bonds) between the
sugars of one nucleotide to the phosphate group of another.
3.3.4 Explain how a DNA double helix is formed (3)
DNA is made of two strands of nucleotides
Nucleotides joined to each other via covalent bonding
The two nucleotide strands are connected together via hydrogen bonds
Nucleotide bases join in a specific pattern; adenine with thymine, cytosine with guanine
Known as complementary base pairing
2 H-bonds between A and T, 3 H-bonds between C and G
3.3.5 Draw an label a simple diagram of the molecular structure of DNA (1)
IB Biology 2013
2
3.4 DNA replication
3.4.1 Explain DNA replication (3)
Double helix unzipped and unwound by enzyme DNA helicase (by breaking H-bonds
between base pairs)
This occurs at the replication fork
Results in two parent strands (strands are separated by helicase)
Free nucleotides in the nucleus pair with the parent strands through complementary base
pairing
Joined together via covalent phosphodiester bonds by DNA polymerase
Double strand reforms into double helix via enzyme
Now have two daughter strands identical to the original strands
3.4.2 Explain the significance of complementary base pairing in the conservation of the base
sequence of DNA (3)
Complementary base pairing ensures that A always pairs with T and C with G
Allows DNA to be copied exactly from each generation to the next conservation of base
sequence of DNA
Prokaryotes allows entire genome to be copied into each generation
Eukaryotes all cells retain exact copy of total genome
3.4.3 State that DNA replication is semi-conservative (1)
DNA replication is semi-conservative
3.5 DNA transcription and translation
3.5.1 Compare the structure of RNA and DNA (3)
DNA RNA
Deoxyribose sugar Ribose sugar
Bases A,C,T,G Bases A,C,U,G
2 polynucleotide strands 1 polynucleotide strand
3.5.2 Outline DNA transcription in terms of formation of complementary RNA strand (2)
1. DNA helix is opened at position of gene
2. Helix unwound by RNA polymerase
3. One of the polynucleotide chains acts as a template strand for mRNA
4. Free RNA nucleotides form complementary strand to DNA through base pairing
5. Phosphodiester bonds on RNA strand formed by RNA polymerase
6. Thymine is replaced by Uracil
IB Biology 2013
3
7. After mRNA is complete the molecule detaches from DNA and leaves nucleus for
cytoplasmic ribosomes
8. DNA helix reforms
3.5.3 Describe the genetic code in terms of codons composed of triplets of bases (2)
Genetic code consists of 4 bases A, T, C, G
A triplet is used to describe group of 3 bases in DNA
A codon is a group of 3 bases in mRNA
Each codon codes for a particular amino acid
There are 64 codons and 20 amino acids
Therefore codons are degenerate (more than 1 codon for a particular amino acid)
Includes 1 start codon (AUG) and 3 stop codons (UGA/UAA/UAG)
Genetic code is universal
3.5.4 Explain the process of translation, leading to polypeptide formation (3)
Translation is process through which proteins are synthesized
Uses ribosomes, mRNA and tRNA
IB Biology 2013
4
1. mRNA is sent out of nucleus to cytoplasm ribosome (composed of rRNA to catalyse reactions)
2. mRNA binds to the small subunit of the ribosome
3. Ribosome moves along the mRNA strand, beginning translation from the start codon (AUG)
4. tRNA (single strand of RNA folded in a particular way, with an acid attachment side and an
anti-codon side) contains anticodons (groups of 3 bases) which are complementary to the
codons on mRNA
5. tRNA will collect the amino acid which corresponds with its anticodon base pairs
6. Then, tRNA binds to the ribosome where its anticodon matches the codon on the mRNA
done through complementary base pairing and form a hydrogen bond
7. 2 tRNA molecules can bind at once.
8. The 2 amino acids carried by the tRNA form a peptide bond
9. The first tRNA then detaches from the ribosome and the second one takes its place the
ribosome moves along the mRNA to the next codon so another tRNA can bind.
10. Process continues until a polypeptide chain is formed.
3.5.5 Discuss the relationship between one gene and one polypeptide (3)
Initially, it was assumed that one gene would code for one polypeptide
Now discovered that the genetic code is universal same bases/codons code for the same
amino acids in all living organisms
Genetic code is degenerate there are 64 codons but only 20 amino acids.
Same amino acid may be coded for by more than 1 codon
Some proteins are composed of a number of polypeptides therefore each polypeptide has
its own gene (e.g. haemoglobin, with 4 polypeptides)
Exceptions:
1. Some genes code or types of RNA which dont produce polypeptides
2. Some genes control the expression of other genes
7.1 DNA structure (HL)
7.1.1 Describe the structure of DNA (2)
DNA has a double helix shape with a uniform diameter along its entire length
Both helices are right-handed
Strand backbones are closer together on one side of the helix called minor groove. Vice
versa = major groove.
IB Biology 2013
5
Phosphate group attached to 5 end of the deoxyribose sugar and hydroxyl (OH) group
attached to 3
Two polynucleotide strands are anti-parallel one strand runs 5 to 3, the other runs 3 to 5
Hydrogen bonding between base pairs purines (A and C) bond to pyramidines (G and T)
7.1.2 Outline the structure of nucleosomes (2)
Nucleosomes consists of DNA wrapped around 8 histone proteins
Held together by another histone protein (H1 linker)
7.1.3 State that nucleosomes help to supercoil chromosomes(1)
Nucleosomes help to super-coil chromosomes and help to regulate transcription
Super coil to factor x 15,000
Super coiled areas cannot be transcribed
Allows control over which genes are expressed
7.1.4 Distinguish between unique and highly repetitive sequences in nuclear DNA (2)
Unique/single-copy genes:
Gene coding region
Approx 1.5% of our DNA codes for a particular polypeptide
3% codes for on/off switches
Highly repetitive sequences:
5 45% of the genome
Once called junk DNA
Non-coding/ not translated
Consists of sequences from 5 300 base pairs long
Used in genetic fingerprinting because they accumulate mutations fast
7.1.5 State that eukaryotic genes can contain exons and introns (1)
Eukaryotic genes can contain exons and introns
IB Biology 2013
6
7.2 DNA replication
7.2.1 State that DNA replication occurs in a 5 3 direction (1)
DNA replication occurs in a 5 3 direction
7.2.2 Explain the process of DNA replication in prokaryotes (3)
Process of DNA replication:
DNA replication occurs during S-phase of interphase
1. Enzyme helicase unzips and unwinds the DNA double helix by breaking apart the hydrogen
bonds between bases into two parent strands
2. Enzyme DNA Polymerase III collects free nucleotides from within the nucleus and creates
two complementary strands
3. Initially they are nucleoside triphosphates (base, sugar with 3 phosphate groups). However,
2 of these phosphate groups break off to provide energy for the synthesis reaction between
base of parent strand and new strand
4. Because DNA replication occurs in a 5 3 direction, will form a leading strand and lagging
strand
Leading strand replicates continuously (same direction as replication fork)
Lagging strand replicates in a leap frog motion (away from replication fork)
In the case of the lagging strand, RNA primase acts as primers and leaves markers
for DNA polymerase III
DNA polymerase III replicates small segments of DNA at a time (known as Okazaki
fragments)
5. DNA polymerase I replaces RNA primers with DNA
6. DNA ligase then joins the Okazaki fragments into one continuous strand
7.2.3 State that DNA replication is initiated at many points in eukaryotic chromosomes (1)
DNA replication is initiated at many points in eukaryotic chromosomes (replication bubbles)
7.3 Transcription
7.3.1 State that transcription is carried out in a 5 3 direction (1)
Transcription is carried out in a 5 3 direction
IB Biology 2013
7
7.3.2 Distinguish between the sense strand and anti-sense strands of DNA (2)
The sense strand is the coding strand with the same base sequence as the transcribed mRNA,
except thymine is replaced by uracil.
The anti-sense strand is the template strand, or non-coding strand which is used for
transcription (making the RNA)
7.3.3 Explain the process of transcription in prokaryotes (3)
Process of transcription in prokaryotes:
Occurs during S phase of interphase in preparation for mitosis/cell division
Occurs in 4 stages:
1. Initiation:
The promoter region allows binding of RNA polymerase, which can then find start
of anti-sense strand, start for transcription and know the direction of transcription
DNA helicase unwinds and unzips DNA, exposing the bases of the anti-sense strand
progressively (in 3 5 direction)
RNA nucleotides form complementary base pairs with anti-sense polynucleotide
strand
Complementary base pairing remain in almost the same order except thymine is
replaced by uracil
2. Elongation
RNA polymerase forms covalent bonds between nucleotides
Energy is released through oxidation of nucleoside triphosphates to synthesize
reaction
Bonds are formed by joining the 5 of a free nucleotide to the 3 of a nucleotide
already part of the mRNA chain
RNA polymerase works along nucleotides until pentose-phosphate backbone is
complete
3. Termination
RNA polymerase reaches the terminator site and the enzyme stops
mRNA is complete
7.3.4 State that eukaryotic RNA needs the removal of introns to form nature mRNA (1)
Eukaryotic RNA needs the removal of introns to form mature mRNA
Before transcription, mRNA processing occurs:
1. Introns are removed (by spliceosomes)
2. Extrons are spliced together
IB Biology 2013
8
3. The mRNA is capped at the ends protects mRNA from being broken down
7.4 - Translation
7.4.1 Explain that each tRNA molecule is recognised by a tRNA-activating enzyme (3)
tRNA molecules have a general clover-leaf structure with three loops
Each tRNA molecule has a different structure and contains double stranded sections (due to
H-bond between base pairs) and loops
Each tRNA molecule is recognized by a tRNA-activating enzyme (aminoacyl-tRNA synthetase)
that binds specific amino acid to the tRNA due to different chemical properties and 3D
structures
Binding process uses ATP for energy
Amino acid binding site at 3 end, with CCA nucleotide sequence
7.4.2 Outline the structure of ribosomes, including protein and RNA composition, large and small
subunits, three tRNA binding sites and mRNA binding sites (2)
IB Biology 2013
9
Ribosomes are made of protein and rRNA (ribosomal RNA)
Consists of small subunit and large subunit
Small subunit: has the mRNA binding site
Large subunit: has 3 sites A, P and E
o A site holds the tRNA carrying next amino acid
o P bonds form; holds tRNA attached to growing polypeptide strand
o E tRNA is discharged
7.4.3 State that translation consists of initiation, elongation, translocation and termination (1)
Translation consists of initiation, elongation, translocation and termination.
Initiation:
Ribosome meets mRNA at MET codon (start codon: AUG)
First tRNA is brought in
Elongation:
Peptide bond is formed between amino acids, forming a polypeptide chain
tRNA binds to codon which corresponds with its anticodon amino acids are added
IB Biology 2013
10
Translocation:
Ribosome releases used tRNA and moves along to the next codon
More tRNA and amino acids are brought in, lengthening the chain
Termination:
Ribosome reaches stop codon
Polypeptide is released
7.4.4 State that translation occurs in a 5 3 direction (1)
Translation occurs in a 5 3 direction
7.4.5 Draw and label a diagram showing the structure of a peptide bond b/w two amino acids (1)
7.4.6 Explain the process of translation, including ribosomes, polysomes, start/stop codons (3)
The process of translation occurs in the following steps:
1. Translation occurs in the cytoplasm ribosomes
2. Ribosomes consists of 2 parts small and large subunits
3. Small subunit binds to the mRNA, whilst large subunit has 3 sites A, P and E
4. tRNA carrying anti-codon UAC for methionine (complementary to start codon AUG) and
occupies the P site
5. A site is free for other complementary tRNA to bind
6. tRNA enters the A site carrying the amino acid according to its anti-codon base sequence,
which is complementary to the codon on the mRNA section it is attached to
7. The amino acids are joined together through peptide bonds (condensation reaction)
IB Biology 2013
11
8. Methionine breaks away energy released allows MET to bond with amino acid on the tRNA
in the A site
9. The used tRNA leaves through the E site
10. More tRNA is brought in and the chain lengthened
11. This process continues until the ribosome reaches a STOP codon
12. At the stop codon, the polypeptide is released
13. The one strand of mRNA can be translated by multiple ribosomes at the same time then
they are known as polysomes
7.4.7 State that free ribosomes synthesise proteins for use primary within the cell(1)
Free ribosomes synthesize proteins for use primarily within the cell
Bound ribosomes (attached to the wall of the rough ER) synthesize proteins primarily for
secretion via vesicles or for lysosomes.
Вам также может понравиться
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryОт EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryРейтинг: 3.5 из 5 звезд3.5/5 (231)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)От EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Рейтинг: 4.5 из 5 звезд4.5/5 (119)
- Never Split the Difference: Negotiating As If Your Life Depended On ItОт EverandNever Split the Difference: Negotiating As If Your Life Depended On ItРейтинг: 4.5 из 5 звезд4.5/5 (838)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaОт EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaРейтинг: 4.5 из 5 звезд4.5/5 (265)
- The Little Book of Hygge: Danish Secrets to Happy LivingОт EverandThe Little Book of Hygge: Danish Secrets to Happy LivingРейтинг: 3.5 из 5 звезд3.5/5 (399)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyОт EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyРейтинг: 3.5 из 5 звезд3.5/5 (2219)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeОт EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeРейтинг: 4 из 5 звезд4/5 (5794)
- Team of Rivals: The Political Genius of Abraham LincolnОт EverandTeam of Rivals: The Political Genius of Abraham LincolnРейтинг: 4.5 из 5 звезд4.5/5 (234)
- The Emperor of All Maladies: A Biography of CancerОт EverandThe Emperor of All Maladies: A Biography of CancerРейтинг: 4.5 из 5 звезд4.5/5 (271)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreОт EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreРейтинг: 4 из 5 звезд4/5 (1090)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersОт EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersРейтинг: 4.5 из 5 звезд4.5/5 (344)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceОт EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceРейтинг: 4 из 5 звезд4/5 (890)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureОт EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureРейтинг: 4.5 из 5 звезд4.5/5 (474)
- The Unwinding: An Inner History of the New AmericaОт EverandThe Unwinding: An Inner History of the New AmericaРейтинг: 4 из 5 звезд4/5 (45)
- The Yellow House: A Memoir (2019 National Book Award Winner)От EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Рейтинг: 4 из 5 звезд4/5 (98)
- High Yield Biochemistry PDFДокумент41 страницаHigh Yield Biochemistry PDFKyle Broflovski100% (2)
- Science: Quarter 3 - Module 4: Central Dogma of Biology: Protein SynthesisДокумент32 страницыScience: Quarter 3 - Module 4: Central Dogma of Biology: Protein SynthesisEricha Solomon56% (9)
- Ricin Purification ProtocolДокумент39 страницRicin Purification Protocolbhavnachawla90100% (1)
- Macromolecule ModelsДокумент3 страницыMacromolecule ModelsBHADUIWAK0% (2)
- Plant Proteins As High-Quality Nutritional Source For Human DietДокумент15 страницPlant Proteins As High-Quality Nutritional Source For Human Dietjoannamae molagaОценок пока нет
- Enzyme Catalysis-Chapter 7 (Part 1)Документ22 страницыEnzyme Catalysis-Chapter 7 (Part 1)OmSilence2651Оценок пока нет
- Preterm LabourДокумент3 страницыPreterm Labourcgao30Оценок пока нет
- Commonwealth Statutory Declaration Form (May 2011) PDFДокумент2 страницыCommonwealth Statutory Declaration Form (May 2011) PDFcgao30Оценок пока нет
- Commonwealth Statutory Declaration Form (May 2011) PDFДокумент2 страницыCommonwealth Statutory Declaration Form (May 2011) PDFcgao30Оценок пока нет
- Theme IV Yr 5d Content Guide - MatrixДокумент9 страницTheme IV Yr 5d Content Guide - Matrixcgao30Оценок пока нет
- Pythagoras WorksheetДокумент2 страницыPythagoras Worksheetcgao30Оценок пока нет
- Student Elective Rotation GuideДокумент3 страницыStudent Elective Rotation Guidecgao30Оценок пока нет
- CD30 Literature Review PlanДокумент1 страницаCD30 Literature Review Plancgao30Оценок пока нет
- CD30 Draft CollationДокумент8 страницCD30 Draft Collationcgao30Оценок пока нет
- CD marker panel guide for flow cytometry cell identificationДокумент2 страницыCD marker panel guide for flow cytometry cell identificationcgao30Оценок пока нет
- Lit Reviews For RX Students v7Документ20 страницLit Reviews For RX Students v7Joshua McdonaldОценок пока нет
- CPPREP4002 - Annotated Unit GuideДокумент8 страницCPPREP4002 - Annotated Unit Guidecgao30Оценок пока нет
- CD30 Literature Review PlanДокумент1 страницаCD30 Literature Review Plancgao30Оценок пока нет
- CD30 Literature Review PlanДокумент1 страницаCD30 Literature Review Plancgao30Оценок пока нет
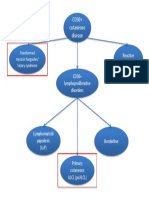
- CD30+ cutaneous lymphoproliferative disordersДокумент1 страницаCD30+ cutaneous lymphoproliferative disorderscgao30Оценок пока нет
- Paediatrics: Acyanotic Heart DiseaseДокумент5 страницPaediatrics: Acyanotic Heart Diseasecgao30Оценок пока нет
- Pelvic Organ ProlapseДокумент5 страницPelvic Organ Prolapsecgao30Оценок пока нет
- Pathology Lecture 2 - NeoplasiaДокумент15 страницPathology Lecture 2 - Neoplasiacgao30Оценок пока нет
- Benign Lesions of The Vulva & Vagina (Table)Документ3 страницыBenign Lesions of The Vulva & Vagina (Table)cgao30Оценок пока нет
- Matrix TopicsДокумент3 страницыMatrix Topicscgao30Оценок пока нет
- EndometriosisДокумент1 страницаEndometriosiscgao30Оценок пока нет
- Fibroids: 1. Red DegenerationДокумент2 страницыFibroids: 1. Red Degenerationcgao30Оценок пока нет
- Pathology Lecture 7 - LiverДокумент11 страницPathology Lecture 7 - Livercgao30Оценок пока нет
- Intro To Motor Systems: (Why Else Would You Go To The Gym, Right?)Документ13 страницIntro To Motor Systems: (Why Else Would You Go To The Gym, Right?)cgao30Оценок пока нет
- O&G GlossaryДокумент3 страницыO&G Glossarycgao30Оценок пока нет
- Skin Terms: DescriptorsДокумент2 страницыSkin Terms: Descriptorscgao30Оценок пока нет
- Pathology Lecture 7 - LiverДокумент11 страницPathology Lecture 7 - Livercgao30Оценок пока нет
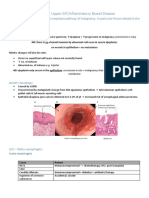
- Pathology Lecture 5 - Upper GITДокумент10 страницPathology Lecture 5 - Upper GITcgao30Оценок пока нет
- Inflammatory Conditions (Disease Mechanisms)Документ3 страницыInflammatory Conditions (Disease Mechanisms)cgao30Оценок пока нет
- Cell Respiration Notes (3&8)Документ9 страницCell Respiration Notes (3&8)cgao30Оценок пока нет
- Understanding Proteins in the Postgenomic Era Through ProteomicsДокумент18 страницUnderstanding Proteins in the Postgenomic Era Through ProteomicsSiddhesh VishwakarmaОценок пока нет
- Module 2: Biochemistry - Amino AcidsДокумент4 страницыModule 2: Biochemistry - Amino AcidsSarah D.Оценок пока нет
- Hepatitis Delta Virus A Fascinating and Neglected Pathogen.Документ11 страницHepatitis Delta Virus A Fascinating and Neglected Pathogen.Diana Carolina di Filippo VillaОценок пока нет
- Intracellular Hepatitis C Virus RNA-dependent RNA Polymerase ActivityДокумент4 страницыIntracellular Hepatitis C Virus RNA-dependent RNA Polymerase ActivitybmnlthyukОценок пока нет
- Protein Metabolism Overview, AnimationДокумент21 страницаProtein Metabolism Overview, AnimationAisyah Arina NurhafizahОценок пока нет
- Assignment On EnzymesДокумент3 страницыAssignment On EnzymesMARK LOUIE SUGANOBОценок пока нет
- Electrophoresis Product GuideДокумент40 страницElectrophoresis Product GuideDolphingОценок пока нет
- 1 s2.0 S0960982214008987 MainДокумент6 страниц1 s2.0 S0960982214008987 MainRia NОценок пока нет
- Protein Structure Hierarchy and RepresentationДокумент31 страницаProtein Structure Hierarchy and RepresentationJayant BhureОценок пока нет
- Biotez Products & Services 2020Документ20 страницBiotez Products & Services 2020skljoleОценок пока нет
- Gen Bio - 1 2.5 - The Organic Molecules of Living OrganismsДокумент7 страницGen Bio - 1 2.5 - The Organic Molecules of Living OrganismsBlaire ReyesОценок пока нет
- MacromoleculesДокумент12 страницMacromoleculesJohn Edward SantosОценок пока нет
- Chapter 4: Amino Acids: MatchingДокумент11 страницChapter 4: Amino Acids: MatchingeliОценок пока нет
- BIO130 Lecture 9Документ14 страницBIO130 Lecture 9lemonpartymanОценок пока нет
- Unit 10 - Mutagenesis PDFДокумент5 страницUnit 10 - Mutagenesis PDFvuivevuiveОценок пока нет
- 1-s2.0-S2666246922000027-Main Expanding The Landscape of E3 Ligases For Targeted Protein DegradationДокумент5 страниц1-s2.0-S2666246922000027-Main Expanding The Landscape of E3 Ligases For Targeted Protein DegradationTsung-Shing WangОценок пока нет
- II TranscriptionДокумент46 страницII TranscriptionGail IbanezОценок пока нет
- General Biology 2: Quarter 3: Module 4Документ4 страницыGeneral Biology 2: Quarter 3: Module 4Ann Lorraine Montealto SadoraОценок пока нет
- Objectives: Terms of Use Privacy Policy Notice AccessibilityДокумент16 страницObjectives: Terms of Use Privacy Policy Notice AccessibilityJuan Sebas OspinaОценок пока нет
- Bio 2-GeneticsДокумент73 страницыBio 2-GeneticsCristina Marie BulloОценок пока нет
- DNA and RNA, Part 2Документ15 страницDNA and RNA, Part 2shiyiОценок пока нет
- DNA Worksheet AnswersДокумент2 страницыDNA Worksheet AnswersJohnric Delacruz50% (2)
- Module 6 RationaleДокумент1 страницаModule 6 RationaleG IОценок пока нет
- Flu Case StudyДокумент4 страницыFlu Case StudyaprilweinmanОценок пока нет