Академический Документы
Профессиональный Документы
Культура Документы
MolBiol 05 DNArepair
Загружено:
Lân LuОригинальное название
Авторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
MolBiol 05 DNArepair
Загружено:
Lân LuАвторское право:
Доступные форматы
MUTATIONS
AND
DNA REPAIR
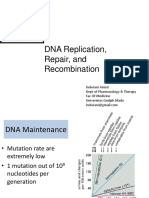
Mutations alter the DNA sequences
Mutations are heritable alterations in the genetic material of any organism
Single strand of normal -globin gene
GTGCACCTGACTCCTGAGGAG
Single strand of mutant -globin gene
GTGCACCTGACTCCTGTGGAG
Single nucleotide change (mutation)
- Single nucleotide change
produces a -globin that
differ from normal globin only by a change
from Glu to Val at the
sixth amino acid position.
- The mutation causes the
disease sickle-cell anemia
The major types of mutation
Base substitution: one base is replaced by another base
Insertion: one or more bases are inserted into the DNA sequence
Deletion: one or more bases are deleted from the DNA sequence
Inversion: a segment of DNA is inverted, but remains at the same overall
location
Duplication: a segment of DNA is duplicated; the second copy usually
remains at the same location as the original
Translocation: a segment of DNA is transfer from its original location to
another position either on the same DNA molecule or on the different DNA
molecule
The nature of mutations
Transitions: pyrimidine-to-pyrimidine and purineto-purine substitutions, such as T to C and A to G
Transversions: pyrimidine-to-purine and purineto-pyrimidine substitutions, such as T to A or G
and A to T or C
- Base substitutions (transitions and transversions), Insertions and Deletions are
simple mutations. Other kinds of mutations might be caused by transposon or
recombination process
- Mutations that alter a single nucleotide are called point mutations
Slippage during DNA replication creates small
insertions or deletions
In human, slippage of trinucleotide (CGG and CAG) repeats causes disease
(fragile X syndrome and Huntingtons disease)
Some Inherited Syndromes with Defects in DNA Repair
Sources of mutation
Inaccuracy in DNA
replication
Chemical damage to
the genetic material
Consequences
1) permanent changes to the DNA (mutation)
2) prevent DNAs use as a template for replication and
transcription
* The cell must scan the genome to detect error in synthesis and
damage to the DNA;
** The cell must mend the lesions do so in that way that, if
possible, restores the original DNA sequence.
REPLICATION ERROR AND
THEIR REPAIR
Proofreading of DNA synthesis
- DNA synthesis: approximately 1 mistake in every 1010 base pair added.
Proofreading allows these mistakes to be corrected.
- DNA polymerase has an error-correcting activity (proofreading exonuclease).
- In palm domain, there are DNA replication site (P) and Exonuclease site (E).
- When a mismatched base pair is detected, the primer:template juntion slides
away from P site and into E site. After the incorrect base pair is removed, the
primer:template junction slides back in to P site and DNA synthesis can continue.
Some replication errors escape proofreading
If the misincorporated nucleotide is not detected and replaced, the sequence
change will become permanent (mutation) in the genome after second round
replication.
DNA mismatch repair system removes errors that
escape proofreading
Mismatch repair pathway for the repair of replication errors
MutS scans the DNA and
detects mismatch
- MutL activates MutH, an enzyme that cause a nick on
one strand near the site of mismatch
-A specific helicase (UrvD) unwinds the DNA, and the
exonuclease progressively digest the mismatch region
- The gap is then filled by DNA polymerase and sealed
with DNA ligase.
Crystal structure of the MutS-DNA complex
How does the cell know which of the two mispaired
bases was the wrong one?
- In E. coli, the parental strand of DNA
is identified by methylation.
-The enzyme Dam methylase converts
Adenine in the sequence GATC to
Methyladenine
- Immediately after DNA replication, the
old strands will be methylated but the
new strands of DNA will not (Hemimethylated)
- The newly synthesized strand is
marked (non-methylated) and is checked
for errors before methylation occurs
Directionality in mismatch repair: exonuclease
removal of mismatched DNA
Different exonucleases are used to remove single stranded DNA between the nick
created by MutH and the mismatch, depending on whether MutH cuts the DNA
on the 5 or 3 side of the misincorporated nucleotide.
Repair mismatch system in eukaryotes
-In eukaryotic cells also repair mismatch and
do so using homologs to MutS (called MSH
proteins for MutS homologs) and MutL
(called MLH and PMS).
- Eukaryotes have multiple MutS-like
proteins with different specificity. For
example, one is specific for simple
mismatch, whereas another recognize small
insertions or deletions resulting from
slippage
- Eukaryotes lack MutH and do not have
hemi-methylation. MSH interacts with the
sliding clamp and recruits mismatch repair
proteins.
DNA DAMAGE
A summary of spontaneous alterations likely to require DNA repair
The sites that are known to be modified by oxidative damage
Hydrolytic attack
Uncontrolled methylation
Depurination and deamination are the most frequent chemical
reaction known to create serious DNA damage in cells
- Depurination: purine bases (A and G) will be lost from DNA
- Deamination: loss of an amino group from cytosine to produce the base uracil
How chemical modifications of nucleotides produce mutations
Deamination
DNA is damaged by alkylation, oxidation
-In alkylation: methyl or ethyl groups are
transferred to reactive sites on the bases and
to phosphate in the DNA backbone
- Oxidation of guanine generates oxoG
(common mutations found in human cancers)
Thymine dimer: UV induces the formation of a cyclobutane ring
between adjacent thymines
These linked bases are incapable of base-pairing and cause the DNA polymerase
to stop during replication
Mutations are also caused by base analogs
Mutations are also caused by intercalating agents
- Intercalating agents are flat molecules containing several polycyclic rings that
bind to the equally flat purine or pyrimidine bases of DNA, just as the bases bind
or stack with each other in the double helix
- Intercalating agents cause the deletion or addition of a base pair or, even few
base pairs
REPAIR OF DNA DAMAGE
Photoreactivation cleaves thymine dimer
In photoreactivation, the enzyme DNA photolyase captures energy from light and
uses it to break the covalent bonds linking adjacent pyrimidine. The damaged
bases are mended directly
Direct reversal of methylated base
The enzyme methyltransferase removes the methyl group from the guanine
residue by transfering it to one of its own Cys residues.
The recognition of an unusual nucleotide in DNA
by base flipping
Structure of a DNA-glycosylase complex
glycosidic
bond
Base excision repair
- An enzyme called glycosylase recognizes
and removes the damaged base by
hydrolyzing the glycosidic bond.
- Glycosylase release the base from the
DNA backbone to leave an AP site (apurinic
or apyrimidic site)
- AP endonuclease and exonuclease cut the
DNA backbone at the 5 and 3 position of
the AP site
- The resulting gap is filled by DNA
polimerase I
- DNA glycosylase are lesion- specific and
cell have multiple DNA glycosylase with
different specificities. A total 8 different
DNA glycosylase have been identified in
human cells.
oxoG:A can be repaired via the base excision pathway
Nucleotide excision repair
UvrC forms a complex
with UvrB and creates
nicks to the 5 and 3 sides
of the lesion
UvrA and UvrB scan the
DNA to identify a
distortion
UvrB melt locally around
the distortion
A comparison of two major DNA repair pathway
Transcription-coupled DNA repair
- RNA polymerase transcribes DNA
normally upstream of the lesion
- Upon encoutering the lesion in DNA,
RNA polymerase stalls and transcription
stops
- RNA polymerase recuits the nucleotide
excision proteins to the site of the lesion
and repairs
- Central to transcription-coupled repair in
eukaryotes is the general transcription
factor TFIIH
Double-strand break (DSB) repair pathway
DSB repair pathway: Non homologous end joining
Three-dimensional structure of a Ku
heterodimer bound to the end of a
duplex DNA fragment
Translesion DNA synthesis enables replication to proceed across
DNA damage
- If cells cannot repair lesions, there is a
fail-safe mechanism that allow the
replication machinery to bypass these
sites of damage. This mechanism is
known as translesion synthesis.
- Translesion synthesis is catalyzed by a
specialized class of DNA polymerase
(UmuC or Y-family DNA polymerase)
DNA repair systems
Вам также может понравиться
- Solution Manual For Genetics From Genes To Genomes 7th Edition Michael Goldberg Janice Fischer Leroy Hood Leland Hartwell DownloadДокумент13 страницSolution Manual For Genetics From Genes To Genomes 7th Edition Michael Goldberg Janice Fischer Leroy Hood Leland Hartwell DownloadTannerBensonkepaq100% (37)
- DNA Damage Repair MechanismДокумент7 страницDNA Damage Repair MechanismSarmistha NayakОценок пока нет
- DNA Mutation and RepairДокумент8 страницDNA Mutation and RepairShaimaa EmadОценок пока нет
- Genetic Material-DNA: 6 November 2003 Reading:The Cell Chapter 5, Pages: 192-201Документ38 страницGenetic Material-DNA: 6 November 2003 Reading:The Cell Chapter 5, Pages: 192-201Laura VgОценок пока нет
- DNA Damage and Tolerance Lecture NotesДокумент5 страницDNA Damage and Tolerance Lecture NotesellieОценок пока нет
- DNA Mutation Workshop GuideДокумент7 страницDNA Mutation Workshop GuideGio GallegoОценок пока нет
- The Replication of DNA: NiveditaДокумент53 страницыThe Replication of DNA: NiveditaLalruatdiki CОценок пока нет
- DNA Damage and RepairДокумент47 страницDNA Damage and RepairdewiulfaОценок пока нет
- Replikasi Dna Repair Dna Rekombinasi Dna: Dr. Upik A. Miskad, PHD Unhas Research CenterДокумент48 страницReplikasi Dna Repair Dna Rekombinasi Dna: Dr. Upik A. Miskad, PHD Unhas Research CenterFitriany LihawaОценок пока нет
- Mutation - Replication InaccuracyДокумент22 страницыMutation - Replication Inaccuracyayeshaabeer874Оценок пока нет
- Genetics and Genomics For Nursing 1st Edition Kenner Test Bank 1Документ36 страницGenetics and Genomics For Nursing 1st Edition Kenner Test Bank 1hollyclarkebfaoejwrny100% (24)
- Repair MechanismДокумент12 страницRepair MechanismrgvendranОценок пока нет
- Dna RepairДокумент34 страницыDna RepairSCRIBEDОценок пока нет
- RizalДокумент25 страницRizalPhia LorenОценок пока нет
- Mrs. Ofelia Solano Saludar: Department of Natural Sciences University of St. La Salle Bacolod CityДокумент57 страницMrs. Ofelia Solano Saludar: Department of Natural Sciences University of St. La Salle Bacolod CitymskikiОценок пока нет
- DNA Rep AirДокумент23 страницыDNA Rep AirTanvir AhmedОценок пока нет
- DNA: Damage Types and Repair Mechanisms (With Diagram)Документ7 страницDNA: Damage Types and Repair Mechanisms (With Diagram)Biofilm NSTUОценок пока нет
- Lecture 5 DNA Mutation and DNA Repair-2Документ65 страницLecture 5 DNA Mutation and DNA Repair-2Constance WongОценок пока нет
- Replication ErrorsДокумент16 страницReplication Errorsilkay turanОценок пока нет
- DNA Replication and RepairДокумент13 страницDNA Replication and RepairViswadeep DasОценок пока нет
- DNA Damage and Mutation Lecture NotesДокумент7 страницDNA Damage and Mutation Lecture NotesellieОценок пока нет
- DNA Damage and RepairДокумент7 страницDNA Damage and RepairAparna AbiОценок пока нет
- DNA Replication, Repair, and RecombinationДокумент114 страницDNA Replication, Repair, and Recombinationviola septinaОценок пока нет
- DNA RepairДокумент23 страницыDNA RepairaliОценок пока нет
- Dna RepairДокумент20 страницDna RepairEaron Van JaboliОценок пока нет
- BCH 405 Lecture 1Документ24 страницыBCH 405 Lecture 1idriscognitoleadsОценок пока нет
- DNA Damage and RepairДокумент25 страницDNA Damage and RepairKhyati GuptaОценок пока нет
- Mbii - l15 - Dna RepairДокумент6 страницMbii - l15 - Dna RepairMiles NsgОценок пока нет
- 6 - Microbial GeneticsДокумент35 страниц6 - Microbial GeneticsAbdallah MoayadОценок пока нет
- 6.1.1 Gene MutationsДокумент43 страницы6.1.1 Gene Mutationsdylanyoung0511Оценок пока нет
- 10-The Mutability & Repair of DNAДокумент62 страницы10-The Mutability & Repair of DNAShoo ShaaОценок пока нет
- Mutations and Dna RepairДокумент18 страницMutations and Dna RepairdrhydrogenОценок пока нет
- Gene Mutation: Genomes 3Документ72 страницыGene Mutation: Genomes 3Abdus SubhanОценок пока нет
- 25.2 DNA Repairv2Документ30 страниц25.2 DNA Repairv2ngguanfangОценок пока нет
- Dna Damage and RepairДокумент23 страницыDna Damage and RepairLathifa FauziaОценок пока нет
- Sw3 Types of Point MutationsДокумент2 страницыSw3 Types of Point MutationsBeaver Sean Felix CaraanОценок пока нет
- MBB425 Lecture 1 - COMPOSITION AND CHEMICAL STRUCTURE OF NUCLEIC ACIDS-Enoch 2019.09.03Документ59 страницMBB425 Lecture 1 - COMPOSITION AND CHEMICAL STRUCTURE OF NUCLEIC ACIDS-Enoch 2019.09.03EMMANUEL ABEL IMAHОценок пока нет
- Dna Genes Chromosomes 2011Документ65 страницDna Genes Chromosomes 2011Suliman GarallehОценок пока нет
- Dna Repair MechanismsДокумент49 страницDna Repair MechanismsayeshaОценок пока нет
- DNA Repair MechanismsДокумент10 страницDNA Repair MechanismsLuke Jovanni TAOCОценок пока нет
- Mutation: Genome Mutations Chromosome Mutations Gene MutationsДокумент65 страницMutation: Genome Mutations Chromosome Mutations Gene MutationsReza WirotamaОценок пока нет
- DNA RepairДокумент6 страницDNA RepairKesana LumangtadОценок пока нет
- DNA Damage and Repair MechanismsДокумент62 страницыDNA Damage and Repair MechanismsnikkiОценок пока нет
- DNA Damage and RepairДокумент32 страницыDNA Damage and RepairNashmiya SaheerОценок пока нет
- L15 DNARepair F2014Документ24 страницыL15 DNARepair F2014jdazuelosОценок пока нет
- DNA Proofreading, EDU161Документ22 страницыDNA Proofreading, EDU161SamanОценок пока нет
- Lecture 26Документ6 страницLecture 26akhilakj117Оценок пока нет
- Dna Repair MechanismsДокумент49 страницDna Repair MechanismsPurushottam GuptaОценок пока нет
- 6.DNA Damages and RepairДокумент49 страниц6.DNA Damages and RepairChandrachur GhoshОценок пока нет
- Dna Mutation & Repair MechanismДокумент23 страницыDna Mutation & Repair MechanismOsama Bin RizwanОценок пока нет
- DNA Damage Repair MechanismsДокумент40 страницDNA Damage Repair MechanismspriannaОценок пока нет
- Unit-6, GeneticsДокумент25 страницUnit-6, GeneticsKerala MekuriyaОценок пока нет
- DNA Repair-RДокумент31 страницаDNA Repair-RAsiah Jelita100% (2)
- The Central Dogma of A Cell: DR S.Balakrishnan, PHD.Документ58 страницThe Central Dogma of A Cell: DR S.Balakrishnan, PHD.joyousОценок пока нет
- DNA Mutation, DNA Repair and Transposable Elements: Micky VincentДокумент33 страницыDNA Mutation, DNA Repair and Transposable Elements: Micky VincentYasmin PrissyОценок пока нет
- Mb17 Dna RepairДокумент19 страницMb17 Dna Repairfaezeh zare karizakОценок пока нет
- 7 DNA DMG and RepairДокумент9 страниц7 DNA DMG and RepairSalaarОценок пока нет
- Chapter 9 - DnaДокумент5 страницChapter 9 - Dnaapi-272677238Оценок пока нет
- DNA RepairДокумент36 страницDNA RepairMeenal MeshramОценок пока нет
- Revision: MODEL 206 (Series 1998 and On) Illustrated Parts CatalogДокумент1 298 страницRevision: MODEL 206 (Series 1998 and On) Illustrated Parts CatalogPedro QAОценок пока нет
- Web Technology Introduction: Cascading Style SheetsДокумент46 страницWeb Technology Introduction: Cascading Style SheetsLân LuОценок пока нет
- 6 ConsiderationsДокумент37 страниц6 ConsiderationsLân LuОценок пока нет
- Web Technology Introduction: Server-Side Script: PHPДокумент21 страницаWeb Technology Introduction: Server-Side Script: PHPLân LuОценок пока нет
- WEBTECH IntroductionДокумент59 страницWEBTECH IntroductionLân LuОценок пока нет
- Web Tech - Course IntroductionДокумент7 страницWeb Tech - Course IntroductionLân LuОценок пока нет
- 4 JavaScriptДокумент66 страниц4 JavaScriptLân LuОценок пока нет
- Topic 2 - Writing A ProposalДокумент18 страницTopic 2 - Writing A ProposalLân LuОценок пока нет
- Exam Molecular BiologyДокумент6 страницExam Molecular BiologyLân Lu100% (2)
- MolBiol RecombinationДокумент45 страницMolBiol RecombinationLân LuОценок пока нет
- Topic 1 - Introduction To Scientific Writing and CommunicationДокумент44 страницыTopic 1 - Introduction To Scientific Writing and CommunicationLân LuОценок пока нет
- MolBiol 08 TranslationДокумент45 страницMolBiol 08 TranslationLân LuОценок пока нет
- MolBiol 07Документ51 страницаMolBiol 07Lân LuОценок пока нет
- BasicConcepts For Assessing Environmental ImpactsДокумент57 страницBasicConcepts For Assessing Environmental ImpactsLân LuОценок пока нет
- CalcI Limits PracticeДокумент18 страницCalcI Limits PracticeLân LuОценок пока нет
- CalcI LimitsДокумент94 страницыCalcI LimitsLân LuОценок пока нет
- MolBiol RegulationДокумент37 страницMolBiol RegulationLân LuОценок пока нет
- MolBiol 07Документ51 страницаMolBiol 07Lân LuОценок пока нет
- MolBiol 02 DNA RNA ChromosomeДокумент70 страницMolBiol 02 DNA RNA ChromosomeLân LuОценок пока нет
- MolBiol 01 Introduction PDFДокумент89 страницMolBiol 01 Introduction PDFLân LuОценок пока нет
- Ylarde vs. Aquino, GR 33722 (DIGEST)Документ1 страницаYlarde vs. Aquino, GR 33722 (DIGEST)Lourdes Loren Cruz67% (3)
- Analysis of Low-Frequency Passive Seismic Attributes in Maroun Oil Field, IranДокумент16 страницAnalysis of Low-Frequency Passive Seismic Attributes in Maroun Oil Field, IranFakhrur NoviantoОценок пока нет
- Man Is Made by His BeliefДокумент2 страницыMan Is Made by His BeliefLisa KireechevaОценок пока нет
- 3er Grado - DMPA 05 - ACTIVIDAD DE COMPRENSION LECTORA - UNIT 2 - CORRECCIONДокумент11 страниц3er Grado - DMPA 05 - ACTIVIDAD DE COMPRENSION LECTORA - UNIT 2 - CORRECCIONANDERSON BRUCE MATIAS DE LA SOTAОценок пока нет
- Oration For Jon Kyle ValdehuezaДокумент2 страницыOration For Jon Kyle ValdehuezaJakes ValОценок пока нет
- Measures-English, Metric, and Equivalents PDFДокумент1 страницаMeasures-English, Metric, and Equivalents PDFluz adolfoОценок пока нет
- A Scenario of Cross-Cultural CommunicationДокумент6 страницA Scenario of Cross-Cultural CommunicationN Karina HakmanОценок пока нет
- Reflection Paper-The Elephant Man PDFДокумент1 страницаReflection Paper-The Elephant Man PDFCarlosJohn02Оценок пока нет
- The Integumentary System Development: Biene, Ellen Angelic Flores, Andrie BonДокумент29 страницThe Integumentary System Development: Biene, Ellen Angelic Flores, Andrie BonMu Lok100% (3)
- COMM 103 Floyd Chapters Study GuideДокумент4 страницыCOMM 103 Floyd Chapters Study GuideMad BasblaОценок пока нет
- Villegas vs. Subido - Case DigestДокумент5 страницVillegas vs. Subido - Case DigestLouvanne Jessa Orzales BesingaОценок пока нет
- Caltech RefДокумент308 страницCaltech RefSukrit ChatterjeeОценок пока нет
- Class NotesДокумент16 страницClass NotesAdam AnwarОценок пока нет
- Air Augmented Rocket (285pages) Propulsion ConceptsДокумент285 страницAir Augmented Rocket (285pages) Propulsion ConceptsAlexandre PereiraОценок пока нет
- Baltimore Catechism No. 2 (Of 4)Документ64 страницыBaltimore Catechism No. 2 (Of 4)gogelОценок пока нет
- Southeast Asian Fabrics and AttireДокумент5 страницSoutheast Asian Fabrics and AttireShmaira Ghulam RejanoОценок пока нет
- Argumentative Essay Project DescriptionДокумент5 страницArgumentative Essay Project DescriptionKaren Jh MoncayoОценок пока нет
- Bug Tracking System AbstractДокумент3 страницыBug Tracking System AbstractTelika Ramu86% (7)
- PED100 Mod2Документ3 страницыPED100 Mod2Risa BarritaОценок пока нет
- Oleracea Contain 13.2% Dry Matter, 15.7% Crude Protein, 5.4% Ether ExtractionДокумент47 страницOleracea Contain 13.2% Dry Matter, 15.7% Crude Protein, 5.4% Ether ExtractionJakin Aia TapanganОценок пока нет
- Endocrine System Unit ExamДокумент3 страницыEndocrine System Unit ExamCHRISTINE JULIANEОценок пока нет
- Amtek Auto Analysis AnuragДокумент4 страницыAmtek Auto Analysis AnuraganuragОценок пока нет
- Public BudgetingДокумент15 страницPublic BudgetingTom Wan Der100% (4)
- Registration - No Candidate Gender Category Rank/Percentage Allotment - Seat Base - Seat Course CollegeДокумент166 страницRegistration - No Candidate Gender Category Rank/Percentage Allotment - Seat Base - Seat Course CollegeCyber ParkОценок пока нет
- Introduction To Sociology ProjectДокумент2 страницыIntroduction To Sociology Projectapi-590915498Оценок пока нет
- Implementation of 7s Framenwork On RestuДокумент36 страницImplementation of 7s Framenwork On RestuMuhammad AtaОценок пока нет
- Osteoporosis: Prepared By: Md. Giash Uddin Lecturer, Dept. of Pharmacy University of ChittagongДокумент30 страницOsteoporosis: Prepared By: Md. Giash Uddin Lecturer, Dept. of Pharmacy University of Chittagongsamiul bashirОценок пока нет
- Articles 62 & 63: Presented By: Muhammad Saad Umar FROM: BS (ACF) - B 2K20Документ10 страницArticles 62 & 63: Presented By: Muhammad Saad Umar FROM: BS (ACF) - B 2K20Muhammad Saad UmarОценок пока нет
- Role of Courts in Granting Bails and Bail Reforms: TH THДокумент1 страницаRole of Courts in Granting Bails and Bail Reforms: TH THSamarth VikramОценок пока нет
- DLL Week 5Документ3 страницыDLL Week 5Nen CampОценок пока нет