Академический Документы
Профессиональный Документы
Культура Документы
Jurnal Lipase Abraham Mora PDF
Загружено:
Abraham Mora TumanggorИсходное описание:
Оригинальное название
Авторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Jurnal Lipase Abraham Mora PDF
Загружено:
Abraham Mora TumanggorАвторское право:
Доступные форматы
ISSN - 0974-2441
Vol 6, Suppl 3, 2013
Research Article
SCREENING SELECTION IDENTIFICATION PRODUCTION AND OPTIMIZATION OF BACTERIAL
LIPASE FROM OIL SPILLED SOIL.
M.VEERAPAGU1, DR .A. SANKARA NARAYANAN1, K.PONMURUGAN2, K.R.JEYA3
1PG Research Department of Microbiology, K.S.R.College of Arts & Science, Tiruchengode, Tamilnadu, India. 2Department of Botany and
Microbiology, College of Sciences, King Saud University, Riyadh, Kingdom of Saudi Arabia. 3Department of Biotechnology, Bharathidasan
University College, Kurumbalur, Perambalur, Tamilnadu, India. Email: mveerapagu@gmail.com
Received: 13 May 2013, Revised and Accepted: 29 may 2013
ABSTRACT
Lipases are glycerol ester hydrolases that catalyze the hydrolysis of triglycerides to free fatty acids and glycerol. Bacterial Lipase producers were
isolated from oil spilled soil from vegetable oil processing factories. One of the twenty isolated strain exhibited a greater zone of clearance than the
others indicating higher lipase activity was selected and identified based on their morphological and physicochemical characteristics and 16s rRNA
sequencing. The effect of incubation time, medium pH, temperature, agitation, inoculums concentration, carbon source and nitrogen source for the
lipase production was studied. The lipase production was maximum at pH7, temperature 370C and incubation time 48 hours by the lipase
producing bacteria BLP2 Pseudomonas gessardii. Increased enzymatic production was obtained when the organisms were cultured in medium
supplemented with 1% protease peptone by Pseudomonas gessardii (168.7Uml-1). The results of the present study demonstrate that the
Pseudomonas gessardii is ideal for extracellular lipase production at industrial level.
Keywords: lipase, Pseudomonas, screening, production, and oil spilled soil.
INTRODUCTION
Lipases are glycerol ester hydrolases that act on acylglycerols to
liberate fatty acids and glycerol. Lipases can hydrolyze long chain
water-insoluble triglycerides into diglycerides, monoglycerides,
glycerol and fatty acids 1,2. Lipases are ubiquitous enzymes which
are widely
Distributed in plants, animals and microbes3. The ability of lipases
to
perform
very
specific
chemical
transformation
(biotransformation) has make them increasingly popular in the food,
detergent, cosmetic, organic synthesis, and pharmaceutical
industries4,5,6.
Lipases are produced by many microorganisms and higher
eukaryotes. Most commercially useful lipases are of microbial origin.
Lipase-producing microorganisms have been found in diverse
habitats such as industrial wastes, vegetable oil processing factories,
dairies, soil contaminated with oil, oilseeds, and decaying food,
compost heaps, coal tips, and hot springs7,8.Lipase producing
microorganisms include bacteria, fungi, yeasts, and actinomyces.
Lipase-producing microorganisms have been found in diverse
habitats such as industrial wastes, vegetable oil processing factories,
dairies, soil contaminated with oil, oilseeds, and decaying food,
compost heaps, coal tips, and hot springs8.
Microbial lipases have gained special industrial attention due to
their stability, selectivity, and broad substrate specificity 9,10.
Microbial enzymes are also more stable than their corresponding
plant and animal enzymes and their production is more convenient
and safer 11 .
Many microorganisms are known as potential producers of
extracellular lipases, including bacteria, yeast, and fungi12. A variety
of extracellular lipases of bacterial origin with different properties
and specificities have been described and characterized.
Extracellular lipase was isolated from many different bacterial
species, including Bacillus13 and Pseudomonas 14,15.
Particular attention is focused on specific classes of enzymes of
species Pseudomonas, that are among the first studied and used in
biotechnological production but also because of their involvement in
bacterial pathogenesis. Lipases are found in a number of
Pseudomonas 16,17,15. Enzymes of P. aeruginosa, P. cepacia and P.
fluorescens obtained in industrial conditions and are used in organic
synthesis, including catalysis of reactions in aqueous solutions18,19.
The bacterial genus Pseudomonas secretes a number of extracellular
enzymes, which include lipases, in response to fluctuating external
nutrients. Interest in Pseudomonas lipases stems either from their
potential usefulness in a variety of biotechnological applications or
from their detrimental effect on stored food products such as
refrigerated milk. The majority of the strains of Pseudomonas sp. are
producers of lipase and phospholipase-C20. A simple and reliable
method for detecting lipase activity in microorganisms has been
described21. This method uses the surfactantTween 80 in a solid
medium to identify a lipolytic activity. The formation of opaque
zones around the colonies is an indication of lipase production by
the organisms. Modifications of this assay use various surfactants in
combination with Nile blue or neets foot oil and Cu2 + salts.
Also, screening of lipase producers on agar plates is frequently done
by using tributyrin as a substrate22 and clear zones around the
colonies indicate production of lipase. Screening systems making use
of chromogenic substrates have also been described23. Plates of a
modified Rhodamine B agar was used to screen lipase activity in a
large number of microorganisms8. Other versions of this method
have been reported24.
Microbial lipases are mostly extracellular and their production is
greatly influenced by medium composition besides physicochemical
factors such as temperature, pH, and dissolved oxygen. The major
factor for the expression of lipase activity has always been reported
as the carbon source, since lipases are inducible enzymes. These
enzymes are generally produced in the presence of a lipid such as
oil or any other inducer, such as triacylglycerols, fatty
acids,hydrolysable esters, Tweens, bile salts, and glycerol 25,26.
However, nitrogen sources and essential micronutrients should also
be carefully considered for growth and production optimization.
Generally, high productivity has been achieved by culture medium
optimization. Optimization of the concentration of each compound
that constitutes a cultivation medium is usually a time-consuming
procedure 5. Microbial lipases are produced mostly by submerged
culture but solid state fermentation methods can be used also. The
use of by-products as substrates for lipase production, adds high
Veerapagu et al.
value and low-cost substrates may reduce the final cost of the
enzyme 27,28. Considering the importance of lipase enzyme, lipase
producing Pseudomonas sp. have been characterized and optimized
in the present study.
MATERIALS AND METHODS
Collection of soil sample
A soil auger was used in collecting soil sample for analysis. The
auger was used to make a depth of 30 cm using a grid or Zig zag
sampling system. The soil sample for physiological analysis was
collected with unused plastic bag sealed with heavy-duty rubber
bounds. All samples were labeled with a permanent waterproof
maker, while the microbiological analyses were collected using 200
ml capacity sterile glass sampling container.
Isolation of lipase producers
The soil samples were collected from different oil mills located at
Dharmapuri and Salem districts enriched by periodic subculturing of
samples in Nutrient Broth (NB) media. They were aseptically
subjected to serial dilutions and plated on Nutrient Agar (NA) and
incubated at 37C for 24, 48 and 72 h. Acceptable plate counts for
bacteria were between 30 300 cfu/ml per plate. After incubation
200 predominant bacterial colonies were isolated and screened for
lipase activity and then subjected to morphological, cultural and
biochemical examinations.
Screening for lipase activity by Tributyrin Clearing Zone (TCZ )
Asian J Pharm Clin Res, Vol 6, Suppl 3, 2013, 62-67
mixture consists of 1ml of crude enzyme, 2.5ml of olive oil was
incubated for 5 minutes. Then the reaction was stopped by adding
1.0ml of 6N HCLand 5ml Benzene. The upper layer 4ml was pipetted
into a test tube and 1.0 ml of cupric acetate pyridine was added. The
FFA dissolved in Benzene was determined by measuring the
absorbency of Benzene solution at 715nm. Lipase activity was
determined by measuring the amount of FFA from the standard
curves of oleic acid. One unit of lipase activity is defined as the
amount of enzyme that liberated 1mol FFA in 1min at 37 C.
Optimization of fermentation conditions
Time course of lipase production
The time course of lipase production was studied in the enzyme
production medium in shake flasks incubated for 60 h. A 5%
inoculum was added to 50 ml of medium, in 500-ml Erlenmeyer
flasks and incubated at 150 rpm on a rotary shaker, at 36C, for 80
hrs. Samples were removed periodically at 8 hr interval and
bacterial growth as well as lipase activity in the culture supernatant
were determined.
Effect of the medium pH and incubation temperature
The effect of pH and temperature of the fermentation medium for
lipase production was performed by varying pH of the medium from
4 to 10 whereas the other parameters were unaltered. For selection
of optimum temperature for the production of lipases, the
temperatures varying from 20 C to 50C were selected by keeping
the remaining parameters same.
The predominant bacteria in the nutrient agatr plate were isolated
and screened for lipolytic activity. Lipolysis is observed directly by
changes in the appearance of the substrate such as tributyrin and
triolein, which are emulsified mechanically in various growth media
and poured into a petri dish. The bacterial isolates were screened for
lipolytic activity on agar plates containing tributyrin (1%, w/v), agar
(2%, w/v) in LuriaBertani medium 29. Lipase production is
indicated by the formation of clear halos around the colonies grown
on tributyrin- containing agar plates 30,31,13.
Effect of Agitation
Characterization of Bacterial lipase producer
Optimum inoculum concentration for lipase enzyme production
was studied by preparing the inoculum as described and varied
inoculums concentration ( 1% to 10 % ) were added to the enzyme
production medium in 500 ml Erlenmeyer flasks containing 100 ml
of liquid medium on a rotary shaker (150 rpm) and incubated at
36C for 24 hrs and the enzyme was assayed.
Halos around the colonies on tributyrin agar plates are considered
as positive colonies for lipase enzyme production. Such colonies are
isolated and identified by phenotypic characterization based on
morphological, biochemical and physiological characters according
to Bergeys Manual of Systematic Bacteriology32.Characterization
and identification of the isolate with higher lipolytic activity was
carried out both biochemically and by 16s r RNA sequencing.
Lipase Enzyme production
The composition of production medium used in this study was:
(%w/v) peptone 0.2; NH4H2PO4 0.1; NaCl 0.25; MgSO47H2O 0.04;
CaCl2.2H2O 0.04; olive oil 2.0 (v/v); pH 7.0; 1-2 drops Tween 80 as
emulsifier. Overnight cultures were suspended in 5ml of sterile
deionised water and used as the inoculum for pre culture to obtain
an initial cell density to adjust the turbidity of 0.5 McFarland
standard. Submerged microbial cultures were incubat-ed in 500 ml
Erlenmeyer flasks containing 100 ml of liquid medium on a rotary
shaker (150 rpm) and incubated at 36C. After 24 hours of
incubation, the culture was centrifuged at 10,000 rpm for 20 min at
4C and the cell free culture supernatant fluid was used as the
sources of extracellular enzyme. The lipase activity in the
supernatant was determined by the colorimetric method. The
bacterial isolate that produced maximum lipase was selected for
further work.
Colorimetric Assay of Lipase Enzyme
method
using Copper Soap
Fatty acids liberated during hydrolysis of an olive oil substrate by
lipase can be determined colorimetrically using a cupric
acetate/pyridine reagent33. Fatty acids complex with copper to
form cupric salts or soaps that absorb light in the visible range
(max 715 nm), yielding a blue color. Quantification of fatty acid
released by lipase is determined by reference to a standard curve
prepared using oleic acid. Olive oil is used a substrate. The reaction
Effect of agitation on lipase production was performed by
incubating the enzyme production medium with inoculated culture
in an orbital shaking incubator at 36C at varying agitation speed
from 110rpm to 200 rpm for 24 hrs. The enzyme was assayed by
colorimetric method after incubation.
Effect of Inoculum concentration
Effect of Carbon source
Effect of carbon source on the lipase production was analysed by
replacing the olive oil with different carbon sources maltose,Glucose,
sucrose, starch, mannitol, Lactose , Fructose, Mannose, arabinose,
galactose at a concentration of (1% w/v) were added into the
production medium in 500 ml Erlenmeyer flasks containing 100 ml
of liquid medium on a rotary shaker (150 rpm) and incubated at
36C for 24 hrs and the enzyme was assayed.
Effect of nitrogen sources
Effect of nitrogen sources on the lipase production was studied by
replacing the nitrogen source with aminoacids alanine, arginine,
asparatic acid, Glutamine, Histidine, leucine, lysine, valine, threonine
and tryptophan, organic nitrogen soures Beef extract, casein,
peptone, proteose peptone, yeast extract, tryptone , meat extract ,
malt extract soyapeptone, soyabean meal and inorganic nitrogen
sources ammonium nitrate, ammonium sulphate ,ammonium
dihydrogen phosphate, ammonium chloride, ammonium acetate ,
ammonium oxalate, calcium nitrate, potassium nitrate, sodium
nitrate and urea at a final concentration of 1 % (w/v ) were added
to the medium and incubated at 36C for 24 hrs in a rotary shaker
(150 rpm).
RESULTS AND DISCUSSION
Isolation and Screening
Lipases are currently used in different industrial products and
processes and new areas of applications are constantly being added,
which include the production of single cell protein, cosmetics,
63
Veerapagu et al.
Asian J Pharm Clin Res, Vol 6, Suppl 3, 2013, 62-67
pulping, lubricants etc34.Lipase producing microbes have been
found in diverse habitats such as industrial wastes, vegetable oil
processing factories, dairies, soil contaminated with oil, oilseeds, and
decaying food , compost heaps, coal tips, and hot springs35.
During the period of research oil spilled soil samples collected from
different oil mills of Dharmapuri and Salem districts were
processed and serially diluted and plated on Nutrient agar medium.
After incubation predominant bacterial colonies were observed. A
total of 200 bacterial colonies were selected and isolated. They were
screened for lipase activity by cultivating them on tributyrin agar
medium and observed for the presence of clear halos
( zone )
around the colonies. Among the 200 isolates 20 shows clear zone in
the tributyrin medium. The bacterial isolate which showed lipolytic
activity was screened for lipase production in the screening medium.
In the screening medium the isolate Pseudomonas gessardii shows
maximum lipase production which produced 12U ml-1 was selected
for further research while others showed less than 5U ml-1.
Identificiation
The bacterial isolate which showed maximum lipase production was
further characterized and identified by morphological, biochemical
characteristics and by 16srRNA sequencing as Pseudomonas
gessardii.
Optimization of fermentation
Time course of lipase production
The incubation time for enzyme production is governed by the
characteristics of the culture and is based on growth rate. In the
present study the production of lipase starts only after 24 hours of
incubation .Lipase was not produced by the Pseduomonas gessardii
at 8 hr and 16 hr of incubation time. The lipase production decreases
after 48 hours. It was reported that maximum lipase activity was
obtained when the physical environment of the fermentation
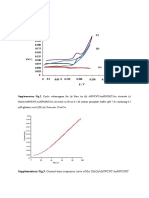
Fig 1: Optimization of incubation time for lipase production
medium was optima for 67 hours for Pseudomonas36. Maximum
lipase production was at 72 hours for Pseudomonas spp37 and
Bacillus coagulans38 and 48 hours for staphylococcus39and
Trichoderma viride 40 respectively.
Effect of the medium pH and incubation temperature
The initial pH of the growth medium influences the rate of lipase
production. It was inferred from the results that the bacteria is
capable of producing lipase from the initial pH of medium from pH
4.0 to pH 10.0. The enzyme production varied considerably from
12.0Uml-1to114Uml-1. The bacteria Pseudomonas gessardii has
optimum lipase production at pH 7.0(114.0U ml-1). However it was
noted that the lipase production was declined with increase in pH
from pH 7.0 to pH 10.0. Maximum lipase activity was observed at
pH7.0 and temperature 37C by staphylococcus 39.
Temperature is a critical parameter that has to be controlled and it
varies from organism to organism. Temperature influences secretion
of extra ceIlular enzymes by changing the physical properties of the
cell membrane.
Fig 2: Optimization of medium pH for lipase production.
Studies conducted for the optimization of temperature shows that
the bacteria produces lipase in wide range of temperature from 20
C to 50C.The lipase enzyme produced at different range of
temperature was from 34.2 U ml-1 to 108.0U ml-1. The optimum
temperature for lipase enzyme production was at 37C
(108.0
U ml-1 ) and the enzyme production was affected and decreased after
increase of temperature above 37C to 50C. It was also noted that
the lipase enzyme production was ceased at temperature
50C.Similar result was reported that the maximum lipase
production was at 37C by Pseudomonas xinjiangensis 41. It was also
reported that the growth and lipase enzyme production of
Pseudomonas fluorescens was maximum at the temperature 360C 42.
Kulkarni and Gadre43 reported that maximum lipase production was
at 250C for Pseudomonas sp.4.
Fig 3: Optimization of Incubation temperature for lipase
production
Effect of Agitation and Inoculum concentration
From the results it was clearly evident that agitation is required for
the bacteria to produce lipase since there was no lipase production
at stationary condition. Agitation at 110 rpm to 160rpm enahanced
the lipase enzyme production .The optimum agitation speed for the
production of lipase by the bacteria was 160rpm (121.6 U ml-1 ). The
rate of agitation speed above 160rpm led to decrease in the enzyme
production. The increase in lipase production could be attributed by
increased oxygen transfer rate, increased surface area of contact
with the media components and better dispersability of the oil
substrate during fermentation under agitated condition. However, at
higher agitation rates, there was a reduction in growth as well as
lipase production 44, 45. Studies conducted for the optimization of
inoculums concentration indicated that the variation in the level of
concentration of inoculum from 1 % to 10 % influence the lipase
enzyme production. The enzyme activity varied from 94.3U ml -1 to
108.0Uml-1. The optimum inoculums concentration for lipase
production was 6%(108.0Uml-1 ).YU Hong-wei et al36. reported that
highest lipase was produced when the inoculums concentration was
6% for Pseudomonas Lip35.
64
Veerapagu et al.
Asian J Pharm Clin Res, Vol 6, Suppl 3, 2013, 62-67
activity49. Similarly inorganic nitrogen sources ammonium sulphate
129.4Uml-1 and ammonium chloride
121.6Uml-1enchanced lipase
production.
Fig.4. Effect of agitation on lipase production by P. gessardii.
Fig. 6.Effect of sugars as additional carbon source on lipase
production by P.gessardii.
Lipase production was very low with urea.
The results of the present study provides useful information for the
optimization of culture conditions such as pH ,temperature,
fermentation time, carbon sources and nitrogen sources to provide
the best lipase production by Pseudomonas species.
Fig. 5 .Effect of Inoculum size on lipase production by
P.gessardii.
These results shows clearly that lipase producing bacteria are
widespread in oil contaminated soil. The optimized growth
conditions developed in this study can be used for a large scale in
industrial purposes.
It is desirable to produce maximum enzyme activity with lower
concentration of inoculums for industrial application.
Effect of carbon source
Lipase production is influenced by the type and concentration of
carbon and nitrogen sources, the culture pH, the growth,
temperature, and the dissolved oxygen concentration 45.Studies
conducted on the effect of sugars supplied as additional carbon
source have not enchaned the lipase production when compared to
control medium. Galactose, maltose and sucrose inhibited the lipase
production. while other sugars tested decreased the level of lipase
enzyme production compared to control medium. It was reported
that starch was the best carbon source for lipase production by P.
fluorescens46. Reduction in the lipase production in the prescence of
sugars as carbon sources could be due to catabolite repression by
readily available carbon sources in the medium14, 36.
Effect of Nitrogen Source
Fig.7.Effect of additional aminoacid as nitrogen source on lipase
production by P.gessardii.
Aminoacid as additional nitrogen source has influenced the lipase
production. Among the 10 aminoacid Histidine ( 135.7 U ml-1 ) and
lysine (126.8 U ml-1 ) has influenced lipase production. On the other
hand asparatic acid, tryptophan, valine, glutamine and alanine
caused a considerable reduction in enzyme production.
On the other hand asparatic acid, tryptophan, valine, glutamine and
alanine caused a considerable reduction in enzyme production. The
effect of additional organic and inorganic nitrogen source 1 % (w/v
) on the production of lipase was studied. Generally, microorganisms
provide high yields of lipase when organic nitrogen sources are
used, such as peptone and yeast extract, which have been used for
lipase production by various thermophilic Bacillus sp. and various
Pseudomonas 47,48. Among the ten different organic nitrogen sources
proteose peptone (168.7 U ml-1 )and peptone (132.5 U ml-1 )
enchanced lipase production whereas lipase production was very
low with casein, soyabean meal and soy peptone. Similar results was
reported for organic nitrogen source by Pseudomonas fluorescens
NS2W41. Among the different nitrogen sources used peptone (2 g/l)
was found to be the most suitable source for maximum lipase
Fig.8.Effect of organic nitrogen source on lipase production by
P.gessardii.
65
Veerapagu et al.
Fig.9.Effect of inorganic nitrogen source on lipase production
by P.gessardii.
REFERENCES
1. Gilham.D. andR.Lehner. Techniques to measure lipase and
esterase activity in vitro. Methods. 2005;36: 139-147.
2. Angkawidjaja, C. and S. Kanaya. Family I.3 lipase: Bacterial
lipases secreted by the type I secretion system. Cell. Mol.
Life Sci. 2006; 63: 2804-2817.
3. Dutta, S. and L. Ray. Production and characterization of an
alkaline thermostable crude lipase from an isolated strain
of Bacillus cereus C7. Appl. Biochem. Biotechnol. 2009; 159:
142-154.
4. Park, H., Lee, K., Chi, Y., & Jeong. S.Effects of methanol on
the catalytic properties of porcine pancreatic lipase. Journal
of Microbiology and Biotechnology. 2005; 15(2): 296301.
5. Gupta, N., Shai, V., & Gupta, R. Alkaline lipase from a novel
strain Burkholderia multivorans: Statistical medium
optimization and production in a bioreactor. Process
Biochemistry. 2007;42(2): 518 526.
6. Grbavcic, S. Z., Dimitrijevic-Brankovic, S. I., Bezbradica, D. I.,
Siler- Marinkovic, S. S., & Knezevic, Z. D. Effect of
fermentation conditions on lipase production by Candida
utilis. Journal of the Serbian Chemical Society. 2007; 72(8
9): 75765.
7. Sztajer H, Maliszewska I, Wieczorek J. Production of
exogenous lipase by bacteria, fungi and actinomycetes.
Enzyme Microb Technol. 1988; 10:492 7.
8. Wang Y, Srivastava KC, Shen GJ, Wang HY. Thermostable
alkaline lipase from a newly isolated thermophilic Bacillus
strain, A30-1 (ATCC 53841). J Ferment Bioeng.1995;79:433
8.
9. Dutra, J. C. V., Terzi, S. C., Bevilaqua, J. V., Damaso, M. C. T.,
Couri,S., Langone, M. A. P. Lipase production in solid state
fermentation monitoring biomass growth of Aspergillus
niger using digital image processing. Applied Biochemistry
and Biotechnology. 2008; 147:6375.
10. Griebeler, N., Polloni, A.E., Remonatto, D., Arbter, F.,
Vardanega, R., Cechet, J.L. et al. Isolation and screening of
lipase producing fungi with hydrolytic activity. Food and
Bioprocess Technology.2011; 4:578-586.
11. Hasan, F., A.A. Shah and A. Hameed. Industrial applications
of microbial lipases. Enzyme Microb. Technol.2006; 39:
235-251.
12. Abada, E. A. E.Production and characterization of a
mesophilic
lipase
isolated
from
Bacillus
stearothermophilus AB-1. Pakistan Journal of Biological
Sciences. 2008; 11: 11001106.
13. Erturul S, Dnmez G, Taka S. Isolation of lipase producing
Bacillus sp. from olive mill wastewater and improving its
enzyme activity. J Hazard Mater 2007;149(3):7204.
14. Kiran GS, Shanmughapriya S, Jayalakshmi J, Selvin J,
Gandhimathi R,Sivaramakrishnan S, Arunkumar M,
Thangavelu T,Natarajaseenivasan K. Optimization of
extracellular psychrophilic alkaline lipase produced by
marine Pseudomonas sp. (MSI057). Bioprocess Biosyst. Eng.
2008; 31: 483-492.
Asian J Pharm Clin Res, Vol 6, Suppl 3, 2013, 62-67
15. Wang S-L, Lin Y-T, Liang T-W, Chio S-H, Ming L-J, Wu P-C.
Purification and characterization of extracellular lipases
from Pseudomonas monteilii TKU009 by the use of soybeans
as the substrate. J. Ind. Microbiol. Biotechnol. 2009; 36: 6573.
16. Kim K, Kwon D, Yoon S, Kim W, Kim K. Purification,
refolding,
and
characterization
of
recombinant
Pseudomonas
fluorescens
lipase.
Protein
Expr.
Purif.2005;39:124-129.
17. Singh S, Banerjee U. Purification and characterization of
trans-3-(4-ethoxyphenyl) glycidic acid methyl ester
hydrolyzing lipase from Pseudomonas aeruginosa. Process.
Biochem. 2007; 42: 1063-1068.
18. Karadzic I, Masui A, Zivkovic L, Fujiwara N. Purification and
characterization of an alkaline lipase from Pseudomonas
aeruginosa isolated from putrid mineral cutting oil as
component
of
metalworking
xuid.J.Biosci.Bioeng.2006;102:82-89.
19. Reetz M, Jaeger K. Overexpression, immobilization and
biotechnological application of Pseudomonas lipases. Chem.
Phys. Lipids. 1998; 93: 3-14.
20. Stoyanova Vasilena, Stanka Georgieva ,Ivan Iliev ,Mariana
Marhova ,Sonya Trifonova. Lipolytic activity of genus
Pseudomonas. J. BioSci. Biotech. 2012; 163-168.
21. Sierra G. A simple method for the detection of lipolytic
activity of microorganisms and some observations on the
influence of the contact between cells and fatty substrates.
Antonie van Leeuwenhoek 1957;23:1522.
22. Cardenas J, Alvarez E, de Castro-Alvarez M-S, SanchezMontero J-M, Valmaseda M, Elson SW, Sinisterra J-V.
Screening and catalytic activity in organic synthesis of
novel fungal and yeast lipases. J Mol Catal B: Enzym.
2001;14:11123
23. Yeoh HH, Wong FM, Lin G. Screening for fungal lipases
using chromogenic lipid substrates. Mycologia. 1986;78:
298300.
24. Kouker G, Jaeger KE. Specific and sensitive plate assay for
bacterial lipases. Appl Environ Microbiol. 1987; 53:2113.
25. Gupta, R., Gupta, N., & Rathi, P. Bacterial lipases: An
overview of production, purification and biochemical
properties. Applied Microbiology and Biotechnology.
2004;64:763781.
26. Sharma, R., Chisti, Y.& Banerjee, U. C.Production,
purification, characterization and applications of lipases.
Biotechnology Advances. 2001;19:627662.
27. Menoncin.S, N.M. Domingues, D.M.G. Freire, G. Toniazzo,
R.L. Cansian and J.V. Oliveira et al., Study of the extraction,
concentration and partial characterization of lipases
obtained from Penicillium verrucosum using solid-state
fermentation of soybean bran. Food Bioprocess Technol.
2010;3:537- 544.
28. Rodriguez, J.A., T. Bhagnagar, S. Roussos, J. Cordova and J.
Baratti et al., Improving lipase production by nutrient
source modification using Rhizopus homthallicus cultured
in solid state fermentation. Process Biochem. 2006; 41:
2264-2269.
29. Fakhreddine L, Kademi A, Ait-Abdelkader N, Baratti JC.
Microbial growth and lipolytic activities of moderate
thermophilic bacterial strains. Biotechnol Lett 1998;20:
87983.
30. Jaeger KE, Ransac S, Dijkstra BW, Colson C, Vanheuvel M,
Misset O. Bacterial lipase.FEMS Microbiol Rev 1994;15:29
63.
31. Kim EK, Jang WH, Ko JH, Kang JS, Noh MJ, Yoo OJ. Lipase
and its modulator from Pseudomonas sp. strain KFCC
10818: proline-to-glutamine substitution at position 112
induces formation of enzymatically active lipase in the
absence of the modulator. J Bacteriol
2001 ;
183(20):59375941.
32. Sneath , P.H.A , Bergeys Manual of Determinative
Bacteriology.2nd ed. Baltimore: Wiliams and Wilkins. (s)
1986: 1105-1138
66
Veerapagu et al.
33. Lowry, R.R. and Tinsley, I.J. Rapid colorimetric
determination of free fatty acids. J. Am. Oil Chem. Soc. 1976;
53:470-472.
34. vijay Gunasekaran and Debabrata Das, Lipase fermentation
: progress and prospects. Indian
Journal of
Biotechnology.2005;4: 437-445.
35. Rohit S, Yusuf C, Ullamchand B. Production,purification,
characterization
and
application
of
lipases.
Biotechnol.Adv.2001;19: 627- 662.
36. YU Hong-wei, HAN Jun, LI Ning, QIE Xiao-sha and JIA Yingmin. Fermentation Performance and Characterization of
Cold-Adapted Lipase Produced with Pseudomonas Lip35.
Agricultural Sciences in China. 2009; 8(8): 956-962.
37. V R Tembhurkar, M B Kulkarni and S A Peshwe.
Optimization of Lipase Production by Pseudomonas spp. in
submerged batch process in shake flask culture . Science
Research Reporter.2012; 2(1):46-50.
38. M.P.PrasanthKumar, A.K.Valsa. Optimization of culture
media and cultural conditions for the production of
extracellular lipase by Bacillus coagulans.
Indian J.
Biotechnol. 2007; 6:114-117 .
39. E. Sirisha ,M. Lakshmi Narasu , and N. Rajasekar. Isolation
and Optimization of Lipase Producing Bacteria from Oil
Contaminated Soils. Advances in Biological Research. 2010;
4 (5): 249-252.
40. M. A.Kashmiri, Ahmad Adnan and Beenish Waseem Butt.
Production, purification and partial characterization of
lipase
from
Trichoderma
Viride.
Afr.
J.
Biotechnol.Vol.2006;5(10): 878-882.
41. KhemikaLomthaisong, Angkhameen Buranarom and
Hataichanoke Niamsup. Investigation of Isolated Lipase
Producing Bacteria from Oil-contaminated Soil with
Asian J Pharm Clin Res, Vol 6, Suppl 3, 2013, 62-67
42.
43.
44.
45.
46.
47.
48.
49.
Proteomic Analysis of its Proteins Responsive to Lipase
Inducer. Journal of BiologicalSciences2012;12:161-167.
N.Kulkarni and RV.Gadre. Production and properties of an
alkaline ,thermophilic lipase from Pseudomonas
fluorescens NS2W. Journal of Industrial Microbiology &
Biotechnology.2002; 28: 344 348.
M. Kavitha and C. Shanthi. Isolation and Characterization of
Cold active lipase producing Pseudomonas sp. 4 from
Marine samples of Tamilnadu Coast. Research Journal of
Biotechnology. 2013; 8 (4) : 57 -62.
Gulati. R, Saxena. R K. and Gupta. R. Fermentation and
downstream processing of lipase from Aspergillus terreus.
Process Biochemistry. 2000; 36:149-155.
E1ibol, M., and Ozer, D. Influence of oxygen transfer on
lipase production by Rhizopus arrhizus. Process
Biochemistry .2001;36: 325-329.
Sztajer H and I Maliszewska. The effect of culture
conditions on lipolytic productivity of microorganisms.
Biotechnol Lett. 1988;10(3 ): 199 204.
Sharma R, Soni SK, Vohra RM, Jolly RS, Gupta LK,Gupta JK.
Production of extracellular alkaline lipase from a Bacillus
sp. RSJ1 and its application in ester hydrolysis. Ind J
Microbiol 2002; 42: 49-54.
Sugihara A, Tani T, Tominaga Y. Purification and .
characterization of a novel thermostable lipase from
Bacillus sp. J. Biochem 1991;109: 211-215.
Kasra-Kermanshahi.R,
Mobarak-Qamsari.E,Moosavinejad.Z. Isolation and Identification of a novel, lipase
producing bacterium, Pseudomonas aeruginosa KM110
Iran.J.M.icrobiol. 2011; 3(2) : 92-98.
67
Вам также может понравиться
- Production and Optimization of Lipase From Candida Rugosa Using Groundnut Oilcake Under Solid State FermentationДокумент7 страницProduction and Optimization of Lipase From Candida Rugosa Using Groundnut Oilcake Under Solid State FermentationInternational Journal of Research in Engineering and TechnologyОценок пока нет
- Actividad EnzimaticДокумент11 страницActividad EnzimaticherfuentesОценок пока нет
- Isolation and Characterization of a Lipase-Producing BacteriumДокумент10 страницIsolation and Characterization of a Lipase-Producing BacteriumRamneet RanaОценок пока нет
- Impo PDFДокумент10 страницImpo PDFRamneet RanaОценок пока нет
- Impo PDFДокумент10 страницImpo PDFRamneet RanaОценок пока нет
- Extraction and Characterization of Lipase Enzymes From Bacillus Cereus (MS6) and Their Medical & Industrial ApplicationsДокумент8 страницExtraction and Characterization of Lipase Enzymes From Bacillus Cereus (MS6) and Their Medical & Industrial ApplicationsInternational Journal of Innovative Science and Research TechnologyОценок пока нет
- Introduction of LipaseДокумент38 страницIntroduction of LipaseRamneet RanaОценок пока нет
- Enzymes Production and ApplicationsДокумент5 страницEnzymes Production and ApplicationsSuraj PatilОценок пока нет
- Optimization of Cellulase Enzyme From Vegetable Waste by Using Trichoderma Atroviride in Solid State FermentationДокумент6 страницOptimization of Cellulase Enzyme From Vegetable Waste by Using Trichoderma Atroviride in Solid State FermentationIOSRjournalОценок пока нет
- LWT - Food Science and TechnologyДокумент6 страницLWT - Food Science and TechnologyIpsita ChakravartyОценок пока нет
- 13 67 Lipase DOtensionДокумент7 страниц13 67 Lipase DOtensionNena Och Exha Part IIОценок пока нет
- MicrobiologyДокумент5 страницMicrobiologygodfrey omariОценок пока нет
- Brazilian Journal of MicrobiologyДокумент7 страницBrazilian Journal of MicrobiologyHimaniОценок пока нет
- Sds-Page Com AzeiteДокумент7 страницSds-Page Com AzeiteInes LealОценок пока нет
- Artigo LipaseДокумент11 страницArtigo LipaseRikson SouzaОценок пока нет
- Lipase Applications in Food IndustryДокумент19 страницLipase Applications in Food IndustryKomagatae Xylinus100% (1)
- 2011 Godoy Et Al Penicillium Simplicissimum Lipase For BiodieselДокумент9 страниц2011 Godoy Et Al Penicillium Simplicissimum Lipase For BiodieseltigapeptindonesiaОценок пока нет
- Microbial Removal of FOGДокумент7 страницMicrobial Removal of FOGIzzat RozaliОценок пока нет
- Experiment 1 - Preparing Nutrient AgarДокумент9 страницExperiment 1 - Preparing Nutrient AgarZuhal AkgunОценок пока нет
- Jurnal: Studi Potensi Lipase Alcaligenes Faecalis Untuk Aplikasi BiodeterjenДокумент6 страницJurnal: Studi Potensi Lipase Alcaligenes Faecalis Untuk Aplikasi BiodeterjenMunib 1Оценок пока нет
- Degradation of Castor Oil and Lipase Production by Pseudomonas AeruginosaДокумент8 страницDegradation of Castor Oil and Lipase Production by Pseudomonas AeruginosaLouella Concepta GoveasОценок пока нет
- MesophilДокумент4 страницыMesophilnurcincelikОценок пока нет
- Production of α-amylase using new strain of Bacillus polymyxa isolated from sweet potatoДокумент7 страницProduction of α-amylase using new strain of Bacillus polymyxa isolated from sweet potatoInternational Organization of Scientific Research (IOSR)Оценок пока нет
- Lipase Production by Geotrichum Candidum-M2Документ5 страницLipase Production by Geotrichum Candidum-M2AparnaОценок пока нет
- Screening of Oleaginous Yeast Strains Tolerant To Lignocellulose Degradation CompoundsДокумент14 страницScreening of Oleaginous Yeast Strains Tolerant To Lignocellulose Degradation CompoundsJuanma AlfaroОценок пока нет
- 2011 Basheer Et Al Aspergillus Awamori Lipase ProductionДокумент12 страниц2011 Basheer Et Al Aspergillus Awamori Lipase ProductiontigapeptindonesiaОценок пока нет
- Apliation AntimicrobianaДокумент4 страницыApliation AntimicrobianaiplabaОценок пока нет
- Lipases in Dairy Industry ReviewДокумент10 страницLipases in Dairy Industry ReviewChinh Đinh ViệtОценок пока нет
- Valorization of Palm Oil Industrial Waste As Feedstock For Lipase ProductionДокумент14 страницValorization of Palm Oil Industrial Waste As Feedstock For Lipase ProductionMangasa SiregarОценок пока нет
- Production and Medium Optimization of Amylase By: Bacillus SPP Using Submerged Fermentation MethodДокумент6 страницProduction and Medium Optimization of Amylase By: Bacillus SPP Using Submerged Fermentation Methodnurul9535Оценок пока нет
- Isolation and Optimization of Lipase Producing Bacteria From Oil Contaminated SoilsДокумент4 страницыIsolation and Optimization of Lipase Producing Bacteria From Oil Contaminated SoilsMuhamad RidzuanОценок пока нет
- Katalis PDFДокумент13 страницKatalis PDFkhansarafidaОценок пока нет
- Research Journal of Pharmaceutical, Biological and Chemical SciencesДокумент7 страницResearch Journal of Pharmaceutical, Biological and Chemical SciencesMaslakhatun NisakdiyahОценок пока нет
- lipase bacterieДокумент27 страницlipase bacteriejmeiiОценок пока нет
- 2 26 1646983092 1ijbtrjun202201Документ10 страниц2 26 1646983092 1ijbtrjun202201TJPRC PublicationsОценок пока нет
- Isolation and Characterization of Starter Culture From Spontaneous Fermentation of SourdoughДокумент4 страницыIsolation and Characterization of Starter Culture From Spontaneous Fermentation of SourdoughHusna AdilaОценок пока нет
- Production of Lipase Using Cassava Peel and Sunflower Oil in Solid-State Fermentation - Preliminary StudyДокумент7 страницProduction of Lipase Using Cassava Peel and Sunflower Oil in Solid-State Fermentation - Preliminary StudyAgriculturaldavidОценок пока нет
- Aspergillus FlavusДокумент6 страницAspergillus FlavusFebri ShaarawyОценок пока нет
- Biocatalysis and Agricultural BiotechnologyДокумент6 страницBiocatalysis and Agricultural BiotechnologydddinzОценок пока нет
- 10 5923 J Food 20140403 04 PDFДокумент6 страниц10 5923 J Food 20140403 04 PDFERIKA MARIE BECERRELОценок пока нет
- Lipase 3Документ9 страницLipase 3Angie Grados LopezОценок пока нет
- Aspergillus TerreusДокумент6 страницAspergillus Terreusgodfrey omariОценок пока нет
- Mic Lipase Production PDFДокумент15 страницMic Lipase Production PDFdahiya76Оценок пока нет
- 25 LSA Jakathi PDFДокумент4 страницы25 LSA Jakathi PDFLife Science ArchivesОценок пока нет
- Lipase Screening 2009Документ6 страницLipase Screening 2009SriArthi100% (2)
- Lactic Acid Production Using Lactic Acid Bacteria Under Optimized ConditionsДокумент6 страницLactic Acid Production Using Lactic Acid Bacteria Under Optimized ConditionsHanan HusseiniОценок пока нет
- A Novel Application of Solid State Culture Production of Lipases byДокумент5 страницA Novel Application of Solid State Culture Production of Lipases byasep muhamadОценок пока нет
- Lipase 1 (PH, Suhu, Stabilitas Properties and Application, Fenomena)Документ5 страницLipase 1 (PH, Suhu, Stabilitas Properties and Application, Fenomena)rozkananda rohmah agustinОценок пока нет
- Moisture in Wheat BranДокумент7 страницMoisture in Wheat BranreddygonaОценок пока нет
- Fermentation and Recovery of L-Glutamic - Acid - FromДокумент8 страницFermentation and Recovery of L-Glutamic - Acid - FromPrachi BhoirОценок пока нет
- Industrial Applications of Microbial LipasesДокумент17 страницIndustrial Applications of Microbial LipasesSonal Manik CОценок пока нет
- A Possible Role of Peptides in The Growth Enhancement of An Industrial Strain of Saccharomyces Sp.Документ8 страницA Possible Role of Peptides in The Growth Enhancement of An Industrial Strain of Saccharomyces Sp.Albert DomingoОценок пока нет
- Research Article Bacillus SubtilisДокумент10 страницResearch Article Bacillus SubtilisDương Nguyễn Thùy DungОценок пока нет
- Adv Biores ShafaasefДокумент6 страницAdv Biores ShafaasefThao ChaungocОценок пока нет
- Activity and Purity of StarterДокумент22 страницыActivity and Purity of StarterRohit PandeyОценок пока нет
- Starter Cultures in Food ProductionОт EverandStarter Cultures in Food ProductionBarbara SperanzaОценок пока нет
- Biofuels: Alternative Feedstocks and Conversion ProcessesОт EverandBiofuels: Alternative Feedstocks and Conversion ProcessesРейтинг: 3 из 5 звезд3/5 (1)
- Polar Lipids: Biology, Chemistry, and TechnologyОт EverandPolar Lipids: Biology, Chemistry, and TechnologyMoghis U. AhmadОценок пока нет
- Microbial Sensing in FermentationОт EverandMicrobial Sensing in FermentationSatinder K. BrarОценок пока нет
- Preparatory Problems for 49th IChOДокумент60 страницPreparatory Problems for 49th IChOCozma TudorОценок пока нет
- Ijaest12 02 03 49 PDFДокумент5 страницIjaest12 02 03 49 PDFAbraham Mora TumanggorОценок пока нет
- Pemanfaatan Media Sosial (Instagram) Sebagai Media Penyajian Kreasi Seni Dalam PembelajaranДокумент13 страницPemanfaatan Media Sosial (Instagram) Sebagai Media Penyajian Kreasi Seni Dalam PembelajaranAbraham Mora TumanggorОценок пока нет
- PEDOT PSS Conductivity Enhancement Through Addition of Imidazolium Ionic LiquidДокумент9 страницPEDOT PSS Conductivity Enhancement Through Addition of Imidazolium Ionic LiquidAbraham Mora TumanggorОценок пока нет
- Ijaest12 02 03 49 PDFДокумент5 страницIjaest12 02 03 49 PDFAbraham Mora TumanggorОценок пока нет
- 637L8 2011Документ81 страница637L8 2011Abraham Mora TumanggorОценок пока нет
- Supplementary Fig.3. Current-Time Responses Curve of The Gluox/Cmwcnt-Aunp/ChitДокумент1 страницаSupplementary Fig.3. Current-Time Responses Curve of The Gluox/Cmwcnt-Aunp/ChitAbraham Mora TumanggorОценок пока нет
- On The Development of Proton Conducting Polymer Membranes For Hydrogen and Methanol Fuel Cells PDFДокумент11 страницOn The Development of Proton Conducting Polymer Membranes For Hydrogen and Methanol Fuel Cells PDFAbraham Mora TumanggorОценок пока нет
- Ijaest12 02 03 49 PDFДокумент5 страницIjaest12 02 03 49 PDFAbraham Mora TumanggorОценок пока нет
- Ijaest12 02 03 49 PDFДокумент5 страницIjaest12 02 03 49 PDFAbraham Mora TumanggorОценок пока нет
- JCPR 2014 6 6 455 459 PDFДокумент5 страницJCPR 2014 6 6 455 459 PDFAbraham Mora TumanggorОценок пока нет
- 23Документ6 страниц23Abraham Mora TumanggorОценок пока нет
- Carbon Nano Tube 5 PDFДокумент4 страницыCarbon Nano Tube 5 PDFAbraham Mora TumanggorОценок пока нет
- A Sensitive and Selective Sensor For Dopamine Determination Based On A Molecularly Imprinted Electropolymer of O-Aminophenol PDFДокумент7 страницA Sensitive and Selective Sensor For Dopamine Determination Based On A Molecularly Imprinted Electropolymer of O-Aminophenol PDFAbraham Mora TumanggorОценок пока нет
- Yamamoto Acupuncture PDFДокумент17 страницYamamoto Acupuncture PDFcarlos100% (1)
- TonsillectomyДокумент27 страницTonsillectomyRho Vince Caño MalagueñoОценок пока нет
- Trauma y Memoria Procedural 2015 Paper Peter LevineДокумент18 страницTrauma y Memoria Procedural 2015 Paper Peter LevineMaría Regina Castro CataldiОценок пока нет
- CSOM of Middle Ear Part 2Документ55 страницCSOM of Middle Ear Part 2Anindya NandiОценок пока нет
- Data Interpret-Medical StudentsДокумент606 страницData Interpret-Medical StudentsIrfan Ali100% (4)
- Case StudyДокумент3 страницыCase StudyAnonymous Gof8o3Оценок пока нет
- hemiplegia دكتور عزونيДокумент4 страницыhemiplegia دكتور عزونيRaed AlhnaityОценок пока нет
- CCДокумент4 страницыCCJaymih Santos AbasoloОценок пока нет
- KIT CD117 A Review On Expression in Normal and Neoplastic TissuesДокумент16 страницKIT CD117 A Review On Expression in Normal and Neoplastic TissuesVlado VladОценок пока нет
- Human Biological and Cultural EvolutionДокумент51 страницаHuman Biological and Cultural Evolutionmichael0202Оценок пока нет
- ACOSTA, Joyce Ara, SBAR 3 (In Good Condition)Документ2 страницыACOSTA, Joyce Ara, SBAR 3 (In Good Condition)Joyce Ara Tumbaga AcostaОценок пока нет
- Endocrine Emergencies CompiledДокумент102 страницыEndocrine Emergencies CompiledSubhkanish RavindraОценок пока нет
- Physiology of Digestive SystemДокумент95 страницPhysiology of Digestive SystemEsha KumavatОценок пока нет
- Psychology: Motivation and EmotionДокумент45 страницPsychology: Motivation and EmotionAisyah AzmiОценок пока нет
- PBL Biochem Lab - EnzymesДокумент4 страницыPBL Biochem Lab - EnzymesGren May Angeli MagsakayОценок пока нет
- Anaphy (Intro - Skeletal System) ReviewerДокумент34 страницыAnaphy (Intro - Skeletal System) ReviewerYanna G.Оценок пока нет
- Investigation of Plasma Protein DisordersДокумент10 страницInvestigation of Plasma Protein DisordersJosiah BimabamОценок пока нет
- Patrick Ch19 p2Документ31 страницаPatrick Ch19 p2NizarAliОценок пока нет
- TRACHEA With TransДокумент56 страницTRACHEA With TransLarry YuloОценок пока нет
- Organs in The Body Quadrants and RegionsДокумент3 страницыOrgans in The Body Quadrants and RegionsDavid HosamОценок пока нет
- Fundamental Unit of Life (STD-IX)Документ6 страницFundamental Unit of Life (STD-IX)Ayushman MohantyОценок пока нет
- Cavum ThoraxДокумент26 страницCavum ThoraxMelsya H UtamiОценок пока нет
- Paget Disease Causes Excessive Bone Formation and DeformationДокумент14 страницPaget Disease Causes Excessive Bone Formation and DeformationOrson SimonОценок пока нет
- Respiratory SystemДокумент26 страницRespiratory SystemYasser AhmedОценок пока нет
- J. Biol. Chem.-1953-Forbes-359-66Документ9 страницJ. Biol. Chem.-1953-Forbes-359-66Fachrun SofiyahОценок пока нет
- 5 SenseДокумент27 страниц5 Sensechici azriniОценок пока нет
- Chapter 43 PosttestДокумент6 страницChapter 43 PosttestGamepage BuckОценок пока нет
- Urinary System For Grade 6Документ2 страницыUrinary System For Grade 6Kent Francis Layaguin75% (4)
- Draft Exam01Документ13 страницDraft Exam01Hannah Pegalan AlegarmeОценок пока нет
- Blood Pressure ChartДокумент9 страницBlood Pressure ChartVer BautistaОценок пока нет