Академический Документы
Профессиональный Документы
Культура Документы
Nead DNA
Загружено:
api-25887855Оригинальное название
Авторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Nead DNA
Загружено:
api-25887855Авторское право:
Доступные форматы
NEWS OF THE WEEK
Briggs and his colleagues sequenced
Across the continent. Entire 16,565 mitochondrial bases extracted from
mitochondrial genomes now bones from Spain, Germany, Croatia, and
exist for these four Neandertal
Russia (see map) and analyzed those genomes
bones and two more, suggesting
low genetic diversity in this along with the one sequenced earlier, which
extinct human. comes from a long bone fragment from Croa-
tia. The bones ranged in age from 38,000 to
Neander Valley 70,000 years old. The team also compared the
ancient DNA with mitochondrial genome
Mezmaiskaya data from about 100 modern individuals.
Vindija Cave
Cave Briggs and postdoc Jeffrey Good found
El Sidrón 55 places out of the 16,565 bases where the
Cave mitochondrial genomes varied across the six
ancient samples. On average, they found
20 differences between any two samples. In
modern humans, about 60 differences exist
between any two samples, making Neander-
Downloaded from www.sciencemag.org on March 27, 2010
ANCIENT DNA tals about one-third as diverse. That isn’t
unexpected given that humans come from
across the globe and that the Neandertals were
Sequencing Neandertal Mitochondrial confined to Europe and Russia, notes John
Relethford, a biological anthropologist at the
Genomes by the Half-Dozen State University of New York College at
Oneonta. The results call into question earlier
Fourteen years ago, sequencing just a few p. 1068), which was announced in February suggestions that Neandertals were divided
hundred bases of mitochondrial DNA from a (Science, 13 February, p. 866). into separate, regional populations, but more
Neandertal drew applause worldwide. Some of that project’s DNA came from data are needed to be sure, says Briggs.
Ancient DNA studies have come a long way mitochondria, and researchers assembled it By the Max Planck group’s calculations,
since then. On page 318, a team led by into the first entire Neandertal mitochon- this diversity translates into the equivalent of
Svante Pääbo of the Max Planck Institute for drial genome in 2008. Sequencing another at most 3500 breeding Neandertal females, or
Evolutionary Anthropology in Leipzig, Ger- mitochondrial genome that way would cost up to 7000 including males, lower than previ-
many, describes a new technique the team as much as $400 million, says Pääbo gradu- ous rough estimates of about 10,000. This
used to decipher the entire mitochondrial ate student Adrian Briggs. That would make so-called effective population size is far less
genomes from five of these extinct humans. comparing multiple Neandertal genomes— than the actual population and represents a
These genomes show relatively little genetic the only way to understand the population theoretical attempt, based on the population’s
diversity among Neandertals scattered size and structure of our closest relative— genetic diversity, to quantify the number of
across Europe and Russia. prohibitively expensive. So Briggs came up individuals who are breeding at any given
“This is an important step for ancient with a better way. His new cost: about $8000 time. For example, Anders Götherström of
DNA,” says Eddy Rubin, whose lab at the per mitochondrial genome. Uppsala University in Sweden calculates that
Lawrence Berkeley National Laboratory in The approach uses probes that recognize although the Swedish population numbers
California did some of the f irst work and isolate only Neandertal mitochondrial 9 million, the effective population size is
sequencing Neandertal nuclear DNA. Adds DNA from all the contaminating DNA in a about 100,000; he estimates that the 3500
Eske Willerslev, who studies ancient DNA at sample. Thus, Briggs winds up sequencing might translate very roughly into about
the University of Copenhagen in Denmark, just the material he’s looking for. 70,000 Neandertals.
the new technique “provides a solution to a To do this, he first attaches short sequence “Low population size may have been a
technical problem: … how to target specific tags to all the pieces of DNA in his sample, general aspect of Neandertal biology,” says
regions of interest.” creating a DNA “library” that can be copied Briggs. With relatively few individuals, the
Ancient DNA is diff icult to sequence to ensure there will be enough ancient DNA species may have been more vulnerable to
because genetic material degrades over time for future use. Using the already-sequenced extinction from climate change or competi-
into small fragments just tens of bases long Neandertal mitochondrial genome as a tem- tion from our ancestors, he adds.
CREDIT: ADRIAN BRIGGS AND JOHANNES KRAUSE
and errors are often introduced into the aging plate, Briggs designed 574 probes to cover Such a small number “is not too surpris-
sequence. Moreover, 99% of fossil DNA tends the entire mitochondrial genome. When a ing,” says ancient DNA expert Alan Cooper
to be contaminating sequence from microbes probe links up with its target sequence in a of the University of Adelaide in Australia, as
and fungi that have infiltrated the decaying mass of DNA fragments, an enzyme copies archaeological evidence had been pointing
bone. Three years ago, researchers began the rest of the DNA in that piece many times toward this. Cooper and others caution that
sequencing all of the DNA in Neandertal sam- over. Thus, Briggs was able to isolate the analyzing mitochondrial DNA has limits as it
ples, then separating out what looked like whole mitochondrial genome and sequence it is “in effect ‘one gene’ because all its genes
human sequence from the mix. New sequenc- using the latest high-throughput sequencing are so tightly linked.” Thus, says Cooper,
ing technologies made the project affordable technologies. Others have used similar “this is just one view of Neandertal evolution-
enough to go after the entire Neandertal approaches, but Briggs tailored his for the ary history.”
nuclear genome (Science, 17 November 2006, short, degraded fragments found in fossils. –ELIZABETH PENNISI
252 17 JULY 2009 VOL 325 SCIENCE www.sciencemag.org
Published by AAAS
Вам также может понравиться
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeОт EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeРейтинг: 4 из 5 звезд4/5 (5794)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreОт EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreРейтинг: 4 из 5 звезд4/5 (1090)
- Never Split the Difference: Negotiating As If Your Life Depended On ItОт EverandNever Split the Difference: Negotiating As If Your Life Depended On ItРейтинг: 4.5 из 5 звезд4.5/5 (838)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceОт EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceРейтинг: 4 из 5 звезд4/5 (895)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersОт EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersРейтинг: 4.5 из 5 звезд4.5/5 (345)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureОт EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureРейтинг: 4.5 из 5 звезд4.5/5 (474)
- The Emperor of All Maladies: A Biography of CancerОт EverandThe Emperor of All Maladies: A Biography of CancerРейтинг: 4.5 из 5 звезд4.5/5 (271)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)От EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Рейтинг: 4.5 из 5 звезд4.5/5 (121)
- The Little Book of Hygge: Danish Secrets to Happy LivingОт EverandThe Little Book of Hygge: Danish Secrets to Happy LivingРейтинг: 3.5 из 5 звезд3.5/5 (400)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyОт EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyРейтинг: 3.5 из 5 звезд3.5/5 (2259)
- The Yellow House: A Memoir (2019 National Book Award Winner)От EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Рейтинг: 4 из 5 звезд4/5 (98)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaОт EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaРейтинг: 4.5 из 5 звезд4.5/5 (266)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryОт EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryРейтинг: 3.5 из 5 звезд3.5/5 (231)
- Team of Rivals: The Political Genius of Abraham LincolnОт EverandTeam of Rivals: The Political Genius of Abraham LincolnРейтинг: 4.5 из 5 звезд4.5/5 (234)
- The Unwinding: An Inner History of the New AmericaОт EverandThe Unwinding: An Inner History of the New AmericaРейтинг: 4 из 5 звезд4/5 (45)
- Reclaimer PDFДокумент8 страницReclaimer PDFSiti NurhidayatiОценок пока нет
- Trainer Manual Internal Quality AuditДокумент32 страницыTrainer Manual Internal Quality AuditMuhammad Erwin Yamashita100% (5)
- Reaction Paper: From The Office of David L. SchutzerДокумент1 страницаReaction Paper: From The Office of David L. Schutzerapi-25887855Оценок пока нет
- Human Culture, An Evolutionary Force: Nicholas WadeДокумент4 страницыHuman Culture, An Evolutionary Force: Nicholas Wadeapi-25887855Оценок пока нет
- Ancient Finger DnaДокумент2 страницыAncient Finger Dnaapi-25887855Оценок пока нет
- Revelations Show King Tut Was Weak, Sickly, But Experts Say It Won't Tarnish Boy-King's ImageДокумент4 страницыRevelations Show King Tut Was Weak, Sickly, But Experts Say It Won't Tarnish Boy-King's Imageapi-25887855Оценок пока нет
- DocsДокумент4 страницыDocsSwastika SharmaОценок пока нет
- sp.1.3.3 Atoms,+Elements+&+Molecules+ActivityДокумент4 страницыsp.1.3.3 Atoms,+Elements+&+Molecules+ActivityBryaniОценок пока нет
- Diffrent Types of MapДокумент3 страницыDiffrent Types of MapIan GamitОценок пока нет
- Main-A5-Booklet (Spreads) PDFДокумент12 страницMain-A5-Booklet (Spreads) PDFanniyahОценок пока нет
- Cet Admissions 2018 FinalДокумент225 страницCet Admissions 2018 FinalkiranОценок пока нет
- BypassGoldManual PDFДокумент6 страницBypassGoldManual PDFBrad FrancОценок пока нет
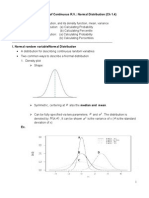
- Distribution of Continuous R.V.: Normal Distribution (CH 1.4) TopicsДокумент7 страницDistribution of Continuous R.V.: Normal Distribution (CH 1.4) TopicsPhạm Ngọc HòaОценок пока нет
- Disbursement VoucherДокумент7 страницDisbursement VoucherDan MarkОценок пока нет
- Normal Consistency of Hydraulic CementДокумент15 страницNormal Consistency of Hydraulic CementApril Lyn SantosОценок пока нет
- Age and Gender Detection Using Deep Learning: HYDERABAD - 501 510Документ11 страницAge and Gender Detection Using Deep Learning: HYDERABAD - 501 510ShyamkumarBannuОценок пока нет
- CM011l E01 Exp.3 DeJuanДокумент3 страницыCM011l E01 Exp.3 DeJuanJarell De JuanОценок пока нет
- Python Cheat Sheet-1Документ8 страницPython Cheat Sheet-1RevathyОценок пока нет
- ESA Mars Research Abstracts Part 2Документ85 страницESA Mars Research Abstracts Part 2daver2tarletonОценок пока нет
- The Ins and Outs Indirect OrvinuДокумент8 страницThe Ins and Outs Indirect OrvinusatishОценок пока нет
- Study On The Form Factor and Full-Scale Ship Resistance Prediction MethodДокумент2 страницыStudy On The Form Factor and Full-Scale Ship Resistance Prediction MethodRaka AdityaОценок пока нет
- PDFДокумент1 страницаPDFJaime Arroyo0% (1)
- 103-Article Text-514-1-10-20190329Документ11 страниц103-Article Text-514-1-10-20190329Elok KurniaОценок пока нет
- LampiranДокумент26 страницLampiranSekar BeningОценок пока нет
- L Rexx PDFДокумент9 страницL Rexx PDFborisg3Оценок пока нет
- Regression Week 2: Multiple Linear Regression Assignment 1: If You Are Using Graphlab CreateДокумент1 страницаRegression Week 2: Multiple Linear Regression Assignment 1: If You Are Using Graphlab CreateSamОценок пока нет
- Solutions Jet FuelДокумент4 страницыSolutions Jet FuelkevinОценок пока нет
- GT-3000 PrintДокумент3 страницыGT-3000 Printmanual imbОценок пока нет
- RRB 17 Sep Set 2 Ibps Guide - Ibps Po, Sbi Clerk, RRB, SSC - Online Mock TestДокумент46 страницRRB 17 Sep Set 2 Ibps Guide - Ibps Po, Sbi Clerk, RRB, SSC - Online Mock TestBharat KumarОценок пока нет
- CompTIA A+ Lesson 3 Understanding, PATA, SATA, SCSIДокумент8 страницCompTIA A+ Lesson 3 Understanding, PATA, SATA, SCSIAli Ghalehban - علی قلعه بانОценок пока нет
- File 1038732040Документ70 страницFile 1038732040Karen Joyce Costales MagtanongОценок пока нет
- Concise Beam DemoДокумент33 страницыConcise Beam DemoluciafmОценок пока нет
- Diagrama Hilux 1KD-2KD PDFДокумент11 страницDiagrama Hilux 1KD-2KD PDFJeni100% (1)
- Module 1 Dynamics of Rigid BodiesДокумент11 страницModule 1 Dynamics of Rigid BodiesBilly Joel DasmariñasОценок пока нет