Академический Документы
Профессиональный Документы
Культура Документы
Multi Density DBScan
Загружено:
Vangelis TaratorisАвторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Multi Density DBScan
Загружено:
Vangelis TaratorisАвторское право:
Доступные форматы
Multi Density DBSCAN
Wesam Ashour and Saad Sunoallah
Islamic University of Gaza, Gaza, Palestine
washour@iugaza.edu.ps, sad.ssa@gmail.com
Abstract. Clustering algorithms are attractive for the task of class identification
in spatial databases. However, the application to large spatial databases rises the
following requirements for clustering algorithms: minimal requirements of
domain knowledge to determine the input parameters, discovery of clusters with
arbitrary shape and good efficiency on large databases.DBSCAN clustering
algorithm relying on a density-based notion of clusters which is designed to
discover clusters of arbitrary shape. DBSCAN requires only one input parameter
and supports the user in determining an appropriate value for it. DBSCAN
cannot find clusters based on difference in densities. We extend the DBSCAN
algorithm so that it can also detect clusters that differ in densities and without the
need to input the value of Eps because our algorithm can find the appropriate
value for each cluster individually by replacing Eps by Local cluster density.
Keywords: Clustering, Arbitrary Shape, DBSCAN, variable Densities.
1 Introduction
The density-based clustering approach is a methodology that is capable of finding
arbitrarily shaped clusters, where clusters are defined as dense regions separated by
low-density regions. A density-based algorithm needs only one scan of the original
data set and can handle noise. The number of clusters is not required, since densitybased clustering algorithms can automatically detect the clusters, along with the
natural number of clusters [1].
Basic density based clustering techniques such as DBSCAN [2] and DENCLUE
[3] treat clusters as regions of high densities separated by regions of no or low
densities. So they are able to suitably handle clusters of different sizes and shapes
besides effectively separating noise (outliers). But they fail to identify clusters with
differing densities unless the clusters are separated by sparse regions.
We extend DBSCAN algorithm to discover clusters with different densities even if
there is no low-density region separates them. Two adjacent spatial regions are
separated into two clusters when the density difference violates a threshold. The
proposed algorithm can automatically find Eps value for each cluster.
1.1 Density Based Clustering (DBSCAN)
DBSCAN is based on the concept of dense areas to form data clustering. The
distribution of points in the cluster should be denser than that outside of the cluster. It
defines a cluster as a maximal set of density-connected points.
H. Yin, W. Wang, and V. Rayward-Smith (Eds.): IDEAL 2011, LNCS 6936, pp. 446453, 2011.
Springer-Verlag Berlin Heidelberg 2011
Multi Density DBSCAN
447
The basic ideas of density-based clustering involve a number of new definition:
The neighborhood within a radius Eps of a given object is called the neighborhood of the object.
If the -neighborhood of an object contains at least a minimum number, MinPts, of
objects, then the object is called a core object.
Given a set of objects, D, we say that an object p is directly density-reachable from
object q if p is within the -neighborhood of q, and q is a core object.
An object p is density-reachable from object q with respect to Eps and MinPts in a
set of objects, D, if there is a chain of objects p1, . . . , pn, where p1 = q and pn = p
such that pi+1 is directly density-reachable from pi with respect to and MinPts, for
1 i n, pi D.
An object p is density-connected to object q with respect to Eps and MinPts in a set
of objects, D, if there is an object O D such that both p and q are densityreachable from O with respect to and MinPts.
Density reachability is the transitive closure of direct density reachability, and this
relationship is asymmetric. Only core objects are mutually density reachable [2].
Fig. 1. Density reachability and density connectivity in density-based clustering (Source: Data
Mining Concepts and Tecniques, Secnd edition, p419)
DBSCAN searches for clusters by checking the -neighborhood of each point in the
database. If the -neighborhood of a point P contains more than MinPts, a new cluster
with P as core point is created. DBSCAN then iteratively collects directly densityreachable points from these core points, which may involve the merge of a few
density-reachable clusters.
2 Related Work
Early efforts like GAM [7] and CLARANS [8] detect clusters within huge data sets
but demand high CPU effort. Later, BIRCH [9] was able to find clusters in the
presence of noise. DBSCAN [2] detect clusters of arbitrary shapes. Recently,
CHAMELEON [10] report clusters of different densities when data sets exhibit
bridges and high discrepancy in density and noise [6].
Another algorithm [4] has introduced a simple idea to improve the results of
DBSCAN algorithm by detecting clusters with large variance in density without
448
W. Ashour and S. Sunoallah
requiring the separation between clusters.This algorithm adds a new point to the
cluster depending on its core point density (the most dense point in the cluster ) which
will cause a problem in nearly similar density neighbor clusters with large internal
density variance, which is solved in our algorithm by calculating the average density
of the cluster in a slow learning process.
Another method [5], DDSC(A Density Differentiated Spatial Clustering
Technique) detects clusters, which are having non-overlapped spatial regions with
reasonable homogeneous density variations within them. If there is significant change
in densities of adjacent regions then all are separated into different clusters.
2.1 DBSCAN Limitation: Sparse Clusters Adjacent to High-Density Clusters
For the data set in Fig. 2, DBSCAN will fall in a trap since it depends on a single Eps
value so it will extract just one cluster from any adjacent clusters.
Fig. 2. Three adjacent clusters with different densities for 4700 point dataset
(a)
(b)
Fig. 3. DBSCAN Result using MinPts=4 and: (a) small Eps value, (b) large Eps value
As we mentioned above DBSCAN depends on single Eps, for small Eps (high
density ) it will extract only the dense class (Fig.3.a) and supposes that other classes
are noise, but for large enough Eps (Fig.3.b) all clusters realize the density conditions
and they are connected so it assumes that they are a single cluster.
Multi Density DBSCAN
449
3 The Proposed Algorithm
As DBSCAN is sensitive to Eps we will use another variable: AVGDST which will
represent the average distances between any point in the cluster and its kth-neighbors,
that variable will be cluster dependent which means it will be varied from a cluster to
another and it will be calculated automatically for each cluster.
In the algorithm we use another concept, since DBSCAN collects the nearest
neighbors using the predefined Eps value, here we calculate DSTp value by taking the
average distance of the nearest kth-neighbors of any point p. The AVGDST of the
cluster and the DSTp will decide if this point belong to that cluster or not.
The algorithm will follow these steps:
1. Calculate the Euclidean distance between each two points of the dataset.
2. For each point p, find the average distance between it and its kth nearest neighbors.
DST p =
qN p
dist ( p, q)
k
Where Np is the group of kth nearest neighbors of p.
3. Insert all points in the queue list.
4. Starting from the most dense point in the queue which have the smallest DST
value and do the following:
(a) Assign the point p to new cluster (Ci).
(b) Remove p from the queue list.
(c) The initial average distances of the cluster will be:
AVGDSTCi = DSTp
(d) Call: gather(Ci, p)
5. gather (Ci, p): For each point q in the nearest kth-neighbors of p do the following:
(a) if (q in queue list ) and ( DSTq var*AVGDSTCi) then:
(i) Add q to Ci members
(ii) Remove q from the queue list.
(iii) Calculate AVGDSTCi from the equation:
AVGDSTi =
AVGDSTi * (N i 1) + DSTq
Ni
(iv) Call: gather(Ci, q)
(v) End if
Where var is the distances variance in the class and its value must be 1, since we
start with the most dense point and the new points added to the cluster will be the
same DST or little larger, this variable will select the density variance in the same
cluster, in other words the new added point to a cluster must has DST value the
same as cluster AVGDST value or with some acceptable difference, this acceptable
450
W. Ashour and S. Sunoallah
difference defined by the variable var. (Ni 1) represent the number of Ci members before adding the new point q.
6. Eliminate very small clusters (noise).
7. Finish.
3.1 K-Value Problems
Since the algorithm depends on k value to calculate the DST for each point, selecting
this value must avoid two problems depending on the dataset we use.
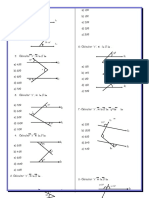
Bridges between two clusters
With huge dataset, some noise points may create bridges between clusters, Fig.4.a,
these bridges lead our algorithm to merge bridged clusters if we use small k value (4
or 5 points), Fig. 4.b, but when increasing the value of k, Fig.4.c, the effect of the
bridge will be eliminated since increasing the k value will increase the Eps value of
the noise faster than the normal points and then the noise will be recognized. In the
other side increasing the value of k leads to high processing time and complexity and
will cause another problem, which is discussed in Fig.5.
(a)
(b)
(c)
Fig. 4. (a) shows two clusters connected by a noise bridge, (b) using k=4, the noise point Eps
value similar to the core points Eps, (c) increasing the value of k, will increase the Eps value of
the noise
Cracks within a cluster
With huge non uniform datasets, cracks within clusters are a normal result, so fixing
the k value to 4 or 5 will cause large clusters with cracks to be separated into two or
more clusters, Fig.5.b, so as the dataset becomes huge the value of k must be
increased to avoid the cracks problem, but a large value of k may cause to merge a
well separated clusters. So the value of k must be large enough to eliminate noise
bridges and connects cluster parts but not too large to merge well separated clusters.
(a)
(b)
(c)
Fig. 5. (a) cracked cluster, (b) choosing k=4 will separate this cluster into two clusters, (c)
increasing the value of k will merge clusters parts
Multi Density DBSCAN
451
4 Experimental Results
Here we evaluate the performance of the proposed algorithm. We implemented this
algorithm in C#. We have used two dimensional datasets to test the algorithm.
(a)
(b)
Fig. 6. (a) 1028 points dataset with four clusters, (b) the result using 2.5 variance, k=15
Fig.6.a shows four clusters of different size, shape, and density. The algorithm
starts from the most dense point ( lies in the green cluster ) and collects the similar
points around it until there is no more similar density around, then it starts from the
most dense point in the remaining points (lies in the red cluster ) and starts over again.
Note that choosing a large enough k value protects from separating the red cluster by
its south crack. The variance value 2.5 chosen by experiments outputs best results.
(a)
(b)
Fig. 7. (a) 1572 points, with 4 clusters and (b) the result using 2.5 variance, k=15
In Fig.7.b we see a slightly different density clusters (red and violet) attached each
other but since the algorithm has no unique Eps value so it separates them easily depending on the density of each cluster. This result may appear using DBSCAN but we
need to select a critical value of Eps to ensure that neighbor clusters (Red and Violet)
will not merge together.
452
W. Ashour and S. Sunoallah
(a)
(b)
Fig. 8. (a) 2972 points, with 10 clusters and (b) the result using 2.5 variance, k=15
In Fig.8.a, we have an artificial data set with 10 clusters differ in shape, size and
density. While there is no Eps value that can help the DBSCAN to find the clusters
due to the variety in densities, our proposed algorithm finds all the clusters successfully as shown in Fig.8.b
In Fig.9 we have three clusters inside each other. DBSCAN will not be able to separate the clusters successfully due the variety in density. If DBSCAN finds the middle
cluster, it will assume that the inner and outer clusters are noise. If we modify Eps in
an attempt to allow DBSCAN to detect the inner and outer clusters successfully, then
DBSCAN will merge all the clusters into one big cluster. Thus there is no value for
Eps that allows DBSCAN to solve this data set. While the correct result will not appear in any Eps value using DBSCAN, the proposed algorithm can find the three clusters easily as shown in Fig. 9.b. These results illuminate the purpose of this algorithm,
where the DBSCAN failed to find the correct clusters.
(a)
(b)
Fig. 9. (a) 4600 points with three clusters and (b) the result using 1.5 variance, k=15
5 Conclusion
In this paper, we have introduced an idea to improve DBSCAN algorithm to solve
two main problems in it, the derivation of Eps value and the problem of connected
Multi Density DBSCAN
453
clusters with different densities since DBSCAN depends on single Eps (threshold)
value to all clusters.
Our algorithm starts from the most dense point as the core of a cluster, that cluster
will grow by gathering similar density neighbor points, when it finishes that cluster it
starts from the most remaining dense point as the core of another cluster an so on. The
algorithm proves itself in many different densities, shapes, sizes, and clusters datasets
with a very good result. In this paper, we show the problems of selecting small value
of k in the large dataset, the noise bridge and the cracked clusters. We show that these
problems can be solved by increasing the value of k.
References
1. Gan, G., Ma, C., Wu, J.: Data Clustering: Theory, Algorithms, and Applications, pp. 67.
SIAM, Philadelphia (2007)
2. Ester, M., Kriegel, H.P., Sander, J., Xu, X.: A density based algorithm for discovering
clusters in large spatial data sets with noise. In: 2nd International Conference on Knowledge Discovery and Data Mining, pp. 226231 (1996)
3. Hinneburg, A., Keim, D.: An efficient approach to clustering in large multimedia data sets
with noise. In: 4th International Conference on Knowledge Discovery and Data Mining,
pp. 5865 (1998)
4. Fahim, A.M., Saake, G., Salem, A.M., Torkey, F.A., Ramadan, M.A.: Improved
DBSCAN for Spatial Databases with Noise and Different Densities. Georgian Electronic
Scientific Journal: Computer Science and Telecommunications, 5360 (2009)
5. Borah, B., Bhattacharyya, D.K.: DDSC: A Density Differentiated Spatial Clustering
Technique. Journal of Computers 3(2), 7279 (2008)
6. Estivill-Castro, V., Lee, I.: AUTOCLUST: Automatic Clustering via Boundary Extraction
for Mining Massive Point-Dta Sets. In: Abrahart, J., Carlisle, B.H. (eds.) Proc. Of the 5th
Int. Conf. on Geocomputation (2000)
7. Openshaw, S.: A Mark 1 Geographical Analysis Machine for the automated analysis of
point data sets. International Journal of GIS 1(4), 335358 (1987)
8. Ng, R.T., Han, J.: Efficient and Effective Clustering Method for Spatial Data Mining.
In: Proceedings of the 20th International Conference on Very Large Data Bases (VLDB),
pp. 144155 (1994)
9. Zhang, T., Ramakrishnan, R., Livny, M.: BIRCH: An Efficient Data Clustering Method
for Very Large Databases. In: Proceedings of the ACM SIGMOD International Conference on Management of Data, pp. 103114 (1996)
10. Karypis, G., Han, E., Kumar, V.: CHAMELEON: A Hierarchical Clustering Algorithm
Using Dynamic Modeling. IEEE Computer: Special Issue on Data Analysis and Mining 32(8), 6875 (1999)
Вам также может понравиться
- M6Документ23 страницыM6foodiegurl2508Оценок пока нет
- DBSCANДокумент8 страницDBSCANvanlxvnitОценок пока нет
- Bde DbscanДокумент11 страницBde DbscanBruno Romero de AzevedoОценок пока нет
- Autoepsdbscan: Dbscan With Eps Automatic For Large Dataset: Manisha Naik Gaonkar & Kedar SawantДокумент6 страницAutoepsdbscan: Dbscan With Eps Automatic For Large Dataset: Manisha Naik Gaonkar & Kedar SawantdjamelОценок пока нет
- Comparison of Density-Based Clustering Algorithms: Mariam RehmanДокумент5 страницComparison of Density-Based Clustering Algorithms: Mariam RehmansuserОценок пока нет
- Data Mining - Density Based ClusteringДокумент8 страницData Mining - Density Based ClusteringRaj EndranОценок пока нет
- Artificial Intelligence: Activity Submitted To: Ms Qurat Ul AinДокумент2 страницыArtificial Intelligence: Activity Submitted To: Ms Qurat Ul AinMisbah Nazeer GondalОценок пока нет
- DS143 Group 13 Presentation-1Документ27 страницDS143 Group 13 Presentation-1Charlton Tatenda UsayiwevhuОценок пока нет
- DBSCAN AlgorithmДокумент15 страницDBSCAN AlgorithmMukesh GautamОценок пока нет
- A Comparative Study of K-Means, DBSCAN and OPTICSДокумент6 страницA Comparative Study of K-Means, DBSCAN and OPTICSkripsi ilyasОценок пока нет
- Data Set Property Based K' in VDBSCAN Clustering AlgorithmДокумент5 страницData Set Property Based K' in VDBSCAN Clustering AlgorithmWorld of Computer Science and Information Technology JournalОценок пока нет
- Sihem JebariДокумент10 страницSihem JebariSihem JebariОценок пока нет
- DBSCANДокумент18 страницDBSCANnilabjyaghoshОценок пока нет
- Ktustudents - In: 1. Hierarchical MethodsДокумент21 страницаKtustudents - In: 1. Hierarchical MethodsE3 TechОценок пока нет
- Chapter 2 (01-09-2019)Документ13 страницChapter 2 (01-09-2019)DerrazОценок пока нет
- BirchДокумент6 страницBirchhehehenotyoursОценок пока нет
- DB ScanДокумент7 страницDB ScanUJJAWAL PUGALIAОценок пока нет
- OpticsДокумент3 страницыOpticshokijic810Оценок пока нет
- CLUSTERING GRID-BASED METHODS Elsayed Hemayed Data Mining CourseДокумент14 страницCLUSTERING GRID-BASED METHODS Elsayed Hemayed Data Mining CourseamjadОценок пока нет
- DT-DBSCAN: Density Based Spatial Clustering in Linear Expected Time Using Delaunay TriangulationДокумент28 страницDT-DBSCAN: Density Based Spatial Clustering in Linear Expected Time Using Delaunay TriangulationJian Park100% (1)
- DIP Lab 13 DBSCAN ClusteringДокумент6 страницDIP Lab 13 DBSCAN Clusteringmahadm.bscs21seecsОценок пока нет
- Density-Based Clustering Algorithm: Presented by - Rohit PaulДокумент12 страницDensity-Based Clustering Algorithm: Presented by - Rohit PaulRohit PaulОценок пока нет
- A Survey of Some Density Based Clustering Techniques PDFДокумент5 страницA Survey of Some Density Based Clustering Techniques PDFsuserОценок пока нет
- An Initial Seed Selection AlgorithmДокумент11 страницAn Initial Seed Selection Algorithmhamzarash090Оценок пока нет
- Kohonen Networks and Clustering: Comparative Performance in Color ClusteringДокумент9 страницKohonen Networks and Clustering: Comparative Performance in Color ClusteringDeybi Ypanaqué SilvaОценок пока нет
- Clustering For BDA - IIДокумент11 страницClustering For BDA - IIhakonaОценок пока нет
- DBSCAN Clustering Algorithm: Presented byДокумент22 страницыDBSCAN Clustering Algorithm: Presented byJarir AhmedОценок пока нет
- 380 389 PDFДокумент10 страниц380 389 PDFAndika SaputraОценок пока нет
- An Empirical Evaluation of Density-Based Clustering TechniquesДокумент8 страницAn Empirical Evaluation of Density-Based Clustering Techniquessuchi87Оценок пока нет
- Chapter - 1: 1.1 OverviewДокумент50 страницChapter - 1: 1.1 Overviewkarthik0484Оценок пока нет
- Ambo University: Inistitute of TechnologyДокумент15 страницAmbo University: Inistitute of TechnologyabayОценок пока нет
- Fast Reciprocal Nearest Neighbors ClusteringДокумент5 страницFast Reciprocal Nearest Neighbors ClusteringVaziel IvansОценок пока нет
- DBSCANДокумент3 страницыDBSCANyevedi5237Оценок пока нет
- DBSCANДокумент42 страницыDBSCANY SAHITHОценок пока нет
- ML Mid2 AnsДокумент24 страницыML Mid2 Ansrayudu shivaОценок пока нет
- Hierarchical Clustering Unit 4 MLДокумент14 страницHierarchical Clustering Unit 4 MLSmriti SharmaОценок пока нет
- Clustering: K-Means, Agglomerative, DBSCAN: Tan, Steinbach, KumarДокумент45 страницClustering: K-Means, Agglomerative, DBSCAN: Tan, Steinbach, Kumarhub23Оценок пока нет
- Unsupervised Learning Clustering IIДокумент17 страницUnsupervised Learning Clustering IIarif.ishaan99Оценок пока нет
- GDBSCANДокумент30 страницGDBSCANkkoushikgmailcomОценок пока нет
- ClusteringДокумент23 страницыClusteringAditya MohiteОценок пока нет
- Jaipur National University: Project Design With SeminarДокумент26 страницJaipur National University: Project Design With SeminarFaizan Shaikh100% (1)
- K - Nearest NeighborДокумент2 страницыK - Nearest NeighborAnshika TyagiОценок пока нет
- K-Nearest Neighbors: KNN Algorithm PseudocodeДокумент2 страницыK-Nearest Neighbors: KNN Algorithm PseudocodeAnshika TyagiОценок пока нет
- S VD For ClusteringДокумент10 страницS VD For ClusteringLMОценок пока нет
- 4 ClusteringДокумент9 страниц4 ClusteringBibek NeupaneОценок пока нет
- Dbsmote: Density-Based Synthetic Minority Over-Sampling TechniqueДокумент21 страницаDbsmote: Density-Based Synthetic Minority Over-Sampling TechniqueIndi Wei-Huan HuОценок пока нет
- Pertemuan-X - Manajemen Data Bagian 2Документ31 страницаPertemuan-X - Manajemen Data Bagian 2Roisyal BarizОценок пока нет
- Unit - 4 DMДокумент24 страницыUnit - 4 DMmintoОценок пока нет
- Applying SR-Tree Technique in DBSCAN Clustering AlgorithmДокумент4 страницыApplying SR-Tree Technique in DBSCAN Clustering AlgorithmInternational Journal of Application or Innovation in Engineering & ManagementОценок пока нет
- DM BS Lec8 ClusteringДокумент48 страницDM BS Lec8 ClusteringrubaОценок пока нет
- 02 Data Mining-Partitioning MethodДокумент8 страниц02 Data Mining-Partitioning MethodRaj EndranОценок пока нет
- Roll Numbers: Bscsf12M054 BSCSF12MO11 Names: Malik-Ubaid-Ul-RAB Adeel-Ul - Rhaman Overview of The Given AssignmentДокумент3 страницыRoll Numbers: Bscsf12M054 BSCSF12MO11 Names: Malik-Ubaid-Ul-RAB Adeel-Ul - Rhaman Overview of The Given AssignmentmalikОценок пока нет
- Parallel Dbscan With Priority R-Tree: Min Chen, Xuedong Gao Huifei LiДокумент4 страницыParallel Dbscan With Priority R-Tree: Min Chen, Xuedong Gao Huifei LiPrashant JhaОценок пока нет
- Unsupervisd Learning AlgorithmДокумент6 страницUnsupervisd Learning AlgorithmShrey DixitОценок пока нет
- Data Mining and Clustering - Benjamin LamДокумент49 страницData Mining and Clustering - Benjamin LamArijit DasОценок пока нет
- DENCLUE 2.0: Fast Clustering Based On Kernel Density EstimationДокумент11 страницDENCLUE 2.0: Fast Clustering Based On Kernel Density EstimationHanif AwalludinОценок пока нет
- 6 Most ImpДокумент17 страниц6 Most Impsuchi87Оценок пока нет
- Path Based Dissimilarity Measured For Thesis Book PreparationДокумент11 страницPath Based Dissimilarity Measured For Thesis Book PreparationKhin MyintОценок пока нет
- Sonnet 18 ParaphraseДокумент1 страницаSonnet 18 ParaphraseDemiОценок пока нет
- Generalized Second Price AuctionДокумент18 страницGeneralized Second Price AuctionVangelis TaratorisОценок пока нет
- MathematicsДокумент6 страницMathematicsVangelis TaratorisОценок пока нет
- Elliptic Curves NotesДокумент2 страницыElliptic Curves NotesVangelis TaratorisОценок пока нет
- Mit 6.857Документ4 страницыMit 6.857Vangelis TaratorisОценок пока нет
- Bms Computer Karang Asem Timur: List Harga LaptopДокумент1 страницаBms Computer Karang Asem Timur: List Harga LaptopBMS COMPUTERОценок пока нет
- Specifications RUNCOДокумент4 страницыSpecifications RUNCOSergio Diaz PereiraОценок пока нет
- Micron Emmc v50Документ26 страницMicron Emmc v50Kausar AliОценок пока нет
- Wiley - VHDL For Logic Synthesis, 3rd Edition - 978-0-470-97797-2Документ2 страницыWiley - VHDL For Logic Synthesis, 3rd Edition - 978-0-470-97797-2Hiten Ahuja0% (1)
- Glitch Works 8085 SBC Rev 3 GW-8085SBC-3: User's Manual and Assembly GuideДокумент17 страницGlitch Works 8085 SBC Rev 3 GW-8085SBC-3: User's Manual and Assembly Guidegord12Оценок пока нет
- Digital Metronome Dm50: Instruction ManualДокумент2 страницыDigital Metronome Dm50: Instruction ManualpepapdОценок пока нет
- AIX SAN BootДокумент4 страницыAIX SAN BootYulin LiuОценок пока нет
- LSC-24 Servo Controller User ManualДокумент19 страницLSC-24 Servo Controller User ManualNicolás BronziniОценок пока нет
- CalcularДокумент5 страницCalcularLuis Alberto Saldarriaga PurizacaОценок пока нет
- ARDUINO BlynkAPP SETUPДокумент13 страницARDUINO BlynkAPP SETUPImran FareedОценок пока нет
- Important QuestionsДокумент49 страницImportant QuestionsBhargav GamitОценок пока нет
- (Bookflare - Net) - Cloud Computing Simply in Depth by Ajit Singh PDFДокумент135 страниц(Bookflare - Net) - Cloud Computing Simply in Depth by Ajit Singh PDFHaftamu Hailu100% (1)
- OMRON PLC-Based Process Control Engineering GuideДокумент228 страницOMRON PLC-Based Process Control Engineering GuideMichael Parohinog GregasОценок пока нет
- Pic16f84a PDFДокумент88 страницPic16f84a PDFBenjamin Michael LandisОценок пока нет
- Video Surveillance Application BrochureДокумент4 страницыVideo Surveillance Application BrochurepeterchanОценок пока нет
- Fujitsu Workplace SystemsДокумент174 страницыFujitsu Workplace Systemsthecorfiot2Оценок пока нет
- Writing Space: The Computer, Hypertext, and The History of Writing Jay David BolterДокумент9 страницWriting Space: The Computer, Hypertext, and The History of Writing Jay David BolterVinaya BhaskaranОценок пока нет
- Introduction To ADT in Eclipse & HANA As Secondary DBДокумент41 страницаIntroduction To ADT in Eclipse & HANA As Secondary DBtopankajsharmaОценок пока нет
- Manually Clearing Print QueueДокумент7 страницManually Clearing Print QueuexcopytechОценок пока нет
- PLC Based ElevatorДокумент10 страницPLC Based ElevatorShisir KandelОценок пока нет
- GR - 19 (Work Planner)Документ25 страницGR - 19 (Work Planner)Deepak ChaudharyОценок пока нет
- You Tube Video CatalogДокумент27 страницYou Tube Video CatalogUma SankarОценок пока нет
- PartitioningДокумент224 страницыPartitioningAnubha Raina DharОценок пока нет
- Design ProblemsДокумент13 страницDesign ProblemsAristotle RiveraОценок пока нет
- A1 PricelistДокумент9 страницA1 PricelistIveta NikolovaОценок пока нет
- 21.1: On-Board Bus Device Driver Pseudocode ExamplesДокумент7 страниц21.1: On-Board Bus Device Driver Pseudocode ExamplesSoundarya SvsОценок пока нет
- Fx-CG10 20 Manager PLUS EДокумент53 страницыFx-CG10 20 Manager PLUS EJ Luis CallasОценок пока нет
- AWAP608HДокумент2 страницыAWAP608HomerbinhussainОценок пока нет
- Parts Manual 35 HP 1977 PDFДокумент39 страницParts Manual 35 HP 1977 PDFziroopОценок пока нет
- Prosafe rsTI32P01J10 01ENДокумент205 страницProsafe rsTI32P01J10 01ENNguyễn QuyếtОценок пока нет