Академический Документы
Профессиональный Документы
Культура Документы
Genes and Phenotypes
Загружено:
John0 оценок0% нашли этот документ полезным (0 голосов)
12 просмотров2 страницыReview of Classical Genetics
Авторское право
© © All Rights Reserved
Доступные форматы
PDF, TXT или читайте онлайн в Scribd
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документReview of Classical Genetics
Авторское право:
© All Rights Reserved
Доступные форматы
Скачайте в формате PDF, TXT или читайте онлайн в Scribd
0 оценок0% нашли этот документ полезным (0 голосов)
12 просмотров2 страницыGenes and Phenotypes
Загружено:
JohnReview of Classical Genetics
Авторское право:
© All Rights Reserved
Доступные форматы
Скачайте в формате PDF, TXT или читайте онлайн в Scribd
Вы находитесь на странице: 1из 2
554
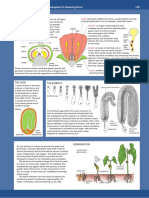
PANEL 81: Review of Classical Genetics
GENES AND PHENOTYPES
Gene:
a functional unit of inheritance, usually corresponding
to the segment of DNA coding for a single protein.
Genome: all of an organisms DNA sequences.
locus: the site of the gene in the genome
Wild-type: the normal,
naturally occurring type
alleles: alternative forms of a gene
homozygous A/A
Mutant: differing from the
wild-type because of a genetic
change (a mutation)
heterozygous a/A
homozygous a/a
GENOTYPE: the specific set of
alleles forming the genome of
an individual
PHENOTYPE: the visible
character of the individual
allele A is dominant (relative to a); allele a is recessive (relative to A)
In the example above, the phenotype of the heterozygote is the same as that of one of the
homozygotes; in cases where it is different from both, the two alleles are said to be co-dominant.
a chromosome at the beginning of the cell
cycle, in G1 phase; the single long bar
represents one long double helix of DNA
CHROMOSOMES
centromere
short p arm
THE HAPLOIDDIPLOID CYCLE
OF SEXUAL REPRODUCTION
long q arm
a chromosome near the end of the cell cycle, in
metaphase; it is duplicated and condensed, consisting of
two identical sister chromatids (each containing one
DNA double helix) joined at the centromere.
short
p arm
long
q arm
pair of
autosomes
maternal 1
paternal 1
paternal 3
maternal 3
paternal 2
maternal 2
Y
X
mother
MEIOSIS
A normal diploid chromosome set, as
seen in a metaphase spread,
prepared by bursting open a cell at
metaphase and staining the scattered
chromosomes. In the example shown
schematically here, there are three
pairs of autosomes (chromosomes
inherited symmetrically from
both parents, regardless of sex) and
two sex chromosomesan X from
the mother and a Y from the father.
The numbers and types of sex
chromosomes and their role in sex
determination are variable from one
class of organisms to another, as is
the number of pairs of autosomes.
HAPLOID
egg
DIPLOID
maternal
chromosome
zygote
A
paternal chromosome
a
diploid germ cell
genotype AB
ab
A
B
MEIOSIS AND
RECOMBINATION
site of crossing-over
genotype aB
a
paternal
chromosome
For simplicity, the cycle is shown for only
one chromosome/chromosome pair.
genotype Ab
maternal chromosome
sperm
SEXUAL FUSION (FERTILIZATION)
sex chromosomes
MEIOSIS AND GENETIC RECOMBINATION
father
DIPLOID
haploid gametes (eggs or sperm)
The greater the distance
between two loci on a single
chromosome, the greater is the
chance that they will be
separated by crossing over
occurring at a site between
them. If two genes are thus
reassorted in x% of gametes,
they are said to be separated on
a chromosome by a genetic map
distance of x map units (or
x centimorgans).
555
TYPES OF MUTATIONS
DELETION: deletes a segment of a chromosome
POINT MUTATION: maps to a single site in the genome,
corresponding to a single nucleotide pair or a very
small part of a single gene
INVERSION: inverts a segment of a chromosome
lethal mutation: causes the developing organism to die
prematurely.
conditional mutation: produces its phenotypic effect only
under certain conditions, called the restrictive conditions.
Under other conditionsthe permissive conditionsthe
effect is not seen. For a temperature-sensitive mutation, the
restrictive condition typically is high temperature, while the
permissive condition is low temperature.
loss-of-function mutation: either reduces or abolishes the
activity of the gene. These are the most common class of
mutations. Loss-of-function mutations are usually
recessivethe organism can usually function normally as
long as it retains at least one normal copy of the affected
gene.
null mutation: a loss-of-function mutation that completely
abolishes the activity of the gene.
TRANSLOCATION: breaks off a segment from one
chromosome and attaches it to another
gain-of-function mutation: increases the activity of the gene
or makes it active in inappropriate circumstances; these
mutations are usually dominant.
dominant-negative mutation: dominant-acting mutation that
blocks gene activity, causing a loss-of-function phenotype
even in the presence of a normal copy of the gene. This
phenomenon occurs when the mutant gene product
interferes with the function of the normal gene product.
suppressor mutation: suppresses the phenotypic effect of
another mutation, so that the double mutant seems normal.
An intragenic suppressor mutation lies within the gene
affected by the first mutation; an extragenic suppressor
mutation lies in a second geneoften one whose product
interacts directly with the product of the first.
TWO GENES OR ONE?
Given two mutations that produce the same phenotype, how can
we tell whether they are mutations in the same gene? If the
mutations are recessive (as they most often are), the answer can
be found by a complementation test.
COMPLEMENTATION:
MUTATIONS IN TWO DIFFERENT GENES
homozygous mutant mother
In the simplest type of complementation test, an individual who
is homozygous for one mutation is mated with an individual
who is homozygous for the other. The phenotype of the
offspring gives the answer to the question.
NONCOMPLEMENTATION:
TWO INDEPENDENT MUTATIONS IN THE SAME GENE
homozygous mutant father
homozygous mutant mother
homozygous mutant father
a1
a2
a1
a2
a1
b
hybrid offspring shows normal phenotype:
one normal copy of each gene is present
a2
hybrid offspring shows mutant phenotype:
no normal copies of the mutated gene are present
Вам также может понравиться
- This Study Resource Was: Drosophila Simulation - Patterns of HeredityДокумент8 страницThis Study Resource Was: Drosophila Simulation - Patterns of HeredityFUN PHYSICS FOR NEET67% (3)
- Unit 3 BiologyДокумент183 страницыUnit 3 BiologyBeverly Crawford75% (8)
- 2 - Modes of InheritanceДокумент42 страницы2 - Modes of InheritanceSimon Grant100% (1)
- Mendelian Genetics and ExtensionsДокумент39 страницMendelian Genetics and ExtensionskcqywОценок пока нет
- HSC Biology NotesДокумент118 страницHSC Biology NotesTanvisha CyrilОценок пока нет
- Genetic Linkage & Mapping (Article) - Khan AcademyДокумент22 страницыGenetic Linkage & Mapping (Article) - Khan Academy嘉雯吳Оценок пока нет
- Principle of Inheritance Variations: Multiple Choice QuestionsДокумент23 страницыPrinciple of Inheritance Variations: Multiple Choice QuestionsUrja Moon100% (1)
- PhylogenyДокумент43 страницыPhylogenyMausam KumravatОценок пока нет
- Brain TeasersДокумент13 страницBrain Teasersshan_batiОценок пока нет
- Di HybridДокумент6 страницDi HybridTaheera Khan100% (2)
- Biology and Ecology of Carp (PDFDrive)Документ389 страницBiology and Ecology of Carp (PDFDrive)Andreea ȘerbanОценок пока нет
- Chemistry 101 Complete NotesДокумент323 страницыChemistry 101 Complete NotesEnerolisa Paredes100% (1)
- M Phase in An Animal CellДокумент2 страницыM Phase in An Animal CellJohnОценок пока нет
- Lesson Plan in ScienceДокумент8 страницLesson Plan in ScienceBALMACEDA DIANA100% (1)
- Mutations: Types and Causes: Mutations Are Recessive or DominantДокумент4 страницыMutations: Types and Causes: Mutations Are Recessive or DominantUsman MaukОценок пока нет
- Cheat Sheet Quiz 1Документ2 страницыCheat Sheet Quiz 1Ryan ThorntonОценок пока нет
- SM Ch05.FinalДокумент22 страницыSM Ch05.Finallgraha13Оценок пока нет
- Linkage and Gene MappingДокумент39 страницLinkage and Gene MappingGilbert ParreñoОценок пока нет
- 2012 Daddy Issues-Paternal Effects On PhenotypeДокумент7 страниц2012 Daddy Issues-Paternal Effects On PhenotypeVishnuGithОценок пока нет
- Linkage and Crossing Over ExplainedДокумент8 страницLinkage and Crossing Over ExplainedMahendra singh RajpurohitОценок пока нет
- Mutation and Genetic Variation: DR - Baraa Maraqa MD General PediatricianДокумент41 страницаMutation and Genetic Variation: DR - Baraa Maraqa MD General Pediatriciananwar jabariОценок пока нет
- Mechanisms of Evolutionary Change in PopulationsДокумент6 страницMechanisms of Evolutionary Change in PopulationsTobias Domenite P.Оценок пока нет
- Dwnload Full Genetics From Genes To Genomes 6th Edition Hartwell Solutions Manual PDFДокумент35 страницDwnload Full Genetics From Genes To Genomes 6th Edition Hartwell Solutions Manual PDFhudsonray4nrf100% (9)
- Full Download Genetics From Genes To Genomes 6th Edition Hartwell Solutions ManualДокумент35 страницFull Download Genetics From Genes To Genomes 6th Edition Hartwell Solutions Manualwoolramisttzdage100% (31)
- Mutations: Types and Causes: Mutations Are Recessive or DominantДокумент4 страницыMutations: Types and Causes: Mutations Are Recessive or DominantIrman FatoniОценок пока нет
- The Ontogeny of PhenotypesДокумент12 страницThe Ontogeny of PhenotypesMarlen PulidoОценок пока нет
- Genetic Disorders TermsДокумент3 страницыGenetic Disorders TermsJohnny eawОценок пока нет
- Introduction To GeneДокумент21 страницаIntroduction To GeneAbhijit NayakОценок пока нет
- 7.13C - Homologs, Orthologs, and Paralogs - Biology LibreTextsДокумент2 страницы7.13C - Homologs, Orthologs, and Paralogs - Biology LibreTextsVIDHI JAINENDRASINH RANAОценок пока нет
- Complementation (Genetics) - Wikipedia, The Free EncyclopediaДокумент4 страницыComplementation (Genetics) - Wikipedia, The Free EncyclopediaShiv KumarОценок пока нет
- Sanders C04Документ38 страницSanders C04Vo UyОценок пока нет
- Basic Concepts of Classical GeneticsДокумент4 страницыBasic Concepts of Classical Geneticsganesandec27Оценок пока нет
- QCB 540 Genetic Basis For VariationДокумент55 страницQCB 540 Genetic Basis For VariationjonathanyewОценок пока нет
- Recombination and LinkageДокумент11 страницRecombination and LinkageHezel DulayОценок пока нет
- 2A GeneticsДокумент47 страниц2A GeneticsYashОценок пока нет
- Gene Expression VarietiesДокумент17 страницGene Expression VarietiesadekОценок пока нет
- Patterns of inheritance in geneticsДокумент13 страницPatterns of inheritance in geneticsALNAKIОценок пока нет
- Gene StructureДокумент5 страницGene StructureLia TuОценок пока нет
- PDF Document-3Документ32 страницыPDF Document-3meryamОценок пока нет
- Cell Differentiation NewДокумент12 страницCell Differentiation NewDiyon JohnОценок пока нет
- AnthropologyДокумент21 страницаAnthropologyAustin WilliamsОценок пока нет
- (Lec3 - Modified Mendelian Dihybrid (Dominance, Lethal GenДокумент18 страниц(Lec3 - Modified Mendelian Dihybrid (Dominance, Lethal Genبلسم محمود شاكرОценок пока нет
- Garvan Institute of Medical Research - Genetics RefreshnerДокумент17 страницGarvan Institute of Medical Research - Genetics RefreshnerLuisaReyesОценок пока нет
- Genetics Canadian 2nd Edition Hartwell Solutions ManualДокумент25 страницGenetics Canadian 2nd Edition Hartwell Solutions ManualCraigVelasquezmcot100% (48)
- Hua Final SolutionsДокумент47 страницHua Final SolutionsbrianhuaОценок пока нет
- Mendelian Genetics ExplainedДокумент53 страницыMendelian Genetics Explainedlaica andalОценок пока нет
- 2025 Module 6 Genetics, Evolution and EcosystemsДокумент42 страницы2025 Module 6 Genetics, Evolution and Ecosystemsharman panwarОценок пока нет
- Chap. 5 Molecular Genetic Techniques: TopicsДокумент28 страницChap. 5 Molecular Genetic Techniques: TopicsanushkaОценок пока нет
- Study Material Unit 2Документ27 страницStudy Material Unit 2api-264649244Оценок пока нет
- The Heritability of Malocclusion: Part 1-Genetics, Principles and TerminologyДокумент11 страницThe Heritability of Malocclusion: Part 1-Genetics, Principles and TerminologyLord JaraxxusОценок пока нет
- ProteomicsДокумент3 страницыProteomicssoumya palavalasaОценок пока нет
- S 2Документ15 страницS 2Feary ProcoratoОценок пока нет
- Sequence HomologyДокумент10 страницSequence HomologyNTA UGC-NETОценок пока нет
- 5 Principles of Inheritance and VariationДокумент7 страниц5 Principles of Inheritance and VariationPreeti AgrawalОценок пока нет
- Imprinting DisordersДокумент19 страницImprinting Disorderssenuberh13Оценок пока нет
- Full Download Genetics Canadian 2nd Edition Hartwell Solutions ManualДокумент35 страницFull Download Genetics Canadian 2nd Edition Hartwell Solutions Manualbeliechopper05srz100% (39)
- Modificationnof Mendelian RatioДокумент26 страницModificationnof Mendelian RatioMonch LacedaОценок пока нет
- Vdocument - in Gene-InteractionДокумент27 страницVdocument - in Gene-InteractionBilal Ahmed KhanОценок пока нет
- Extending Mendelian Genetics For Two or More GenesДокумент27 страницExtending Mendelian Genetics For Two or More GenesGerald GuevarraОценок пока нет
- Definition of Gene (Repaired)Документ107 страницDefinition of Gene (Repaired)Ananda PreethiОценок пока нет
- Genetic Basis For VariationДокумент4 страницыGenetic Basis For VariationNicholas OwОценок пока нет
- Principles of GeneticsДокумент4 страницыPrinciples of GeneticsLucyellowOttemoesoeОценок пока нет
- POG Lecture 12Документ7 страницPOG Lecture 12meilunlyОценок пока нет
- Lecture 6Документ21 страницаLecture 6Sharmitha SaravananОценок пока нет
- Submitted By: Jochelle M. Santos 8-Porifera Submitted To: Ms. Maelyn CamposanoДокумент28 страницSubmitted By: Jochelle M. Santos 8-Porifera Submitted To: Ms. Maelyn CamposanoRichelle SantosОценок пока нет
- Module 3 Mendelian GeneticsДокумент14 страницModule 3 Mendelian GeneticsRaiza AwatОценок пока нет
- ChimieДокумент10 страницChimieDana CapbunОценок пока нет
- MIT3 091SCF09 SummaryДокумент33 страницыMIT3 091SCF09 SummarydavidraaamosОценок пока нет
- Quimica Organica IIДокумент18 страницQuimica Organica IIvskywokervsОценок пока нет
- 508 Lecture 12Документ3 страницы508 Lecture 12JohnОценок пока нет
- AP Chem NotesДокумент45 страницAP Chem NotesSajiveSivalingamОценок пока нет
- Open Educational Resources Textbook ListДокумент35 страницOpen Educational Resources Textbook ListJohn100% (1)
- The Stock Market Jumpstarter V2.0Документ51 страницаThe Stock Market Jumpstarter V2.0Dennis LandichoОценок пока нет
- Semicond LasersДокумент23 страницыSemicond LasersurimОценок пока нет
- Spie Trial ManualДокумент9 страницSpie Trial ManualJohnОценок пока нет
- IPLS Report 2015 v2Документ40 страницIPLS Report 2015 v2JohnОценок пока нет
- Statics SlidesДокумент35 страницStatics SlidesHabibullah RadziОценок пока нет
- X Ray Diffraction 1057Документ5 страницX Ray Diffraction 1057JohnОценок пока нет
- AcДокумент44 страницыAcJohnОценок пока нет
- Thermodynamics: Course IntroductionДокумент52 страницыThermodynamics: Course IntroductionRodrigo Silveira da SilveiraОценок пока нет
- The Effect of Vibrational Excitation of Molecules Involving Methane & NitrogenДокумент15 страницThe Effect of Vibrational Excitation of Molecules Involving Methane & NitrogenRay FraustoОценок пока нет
- Redox PotentialsДокумент1 страницаRedox PotentialsJohnОценок пока нет
- XML For Software Engineers: Tutorial OutlineДокумент47 страницXML For Software Engineers: Tutorial OutlineJohnОценок пока нет
- Flower DevelopmentДокумент1 страницаFlower DevelopmentJohnОценок пока нет
- Exponential Notation and Physical QuantitiesДокумент4 страницыExponential Notation and Physical QuantitiesJohnОценок пока нет
- Fourier Problem and AnswerДокумент2 страницыFourier Problem and AnswerJohnОценок пока нет
- Actin Filaments: 968 PANEL 16-1: The Three Major Types of Protein Filaments That Form The CytoskeletonДокумент1 страницаActin Filaments: 968 PANEL 16-1: The Three Major Types of Protein Filaments That Form The CytoskeletonJohnОценок пока нет
- Molecular MechanismsДокумент2 страницыMolecular MechanismsJohnОценок пока нет
- Plant Tissue SystemsДокумент2 страницыPlant Tissue SystemsJohn0% (1)
- PolymerizationДокумент2 страницыPolymerizationJohnОценок пока нет
- Actin Filaments and MicrotubulesДокумент2 страницыActin Filaments and MicrotubulesJohnОценок пока нет
- Signal Sequences and Protein TranslocationДокумент1 страницаSignal Sequences and Protein TranslocationJohnОценок пока нет
- Panel 11-2Документ1 страницаPanel 11-2nrf2Оценок пока нет
- Squid Giant AxonДокумент1 страницаSquid Giant AxonJohnОценок пока нет
- Matters of Sex LongДокумент3 страницыMatters of Sex Longjenifer lolaОценок пока нет
- Learning Modules For General Biology 2Документ12 страницLearning Modules For General Biology 2Albert Rosete0% (1)
- Co DominanceДокумент24 страницыCo Dominancelucky mayОценок пока нет
- Science Notebook Mendelian GeneticsДокумент5 страницScience Notebook Mendelian GeneticsBasma AhmedОценок пока нет
- Sex-Linked Traits Worksheet: Background InformationДокумент4 страницыSex-Linked Traits Worksheet: Background InformationWilson RobertsОценок пока нет
- SEX DETERMINATION MECHANISMSДокумент33 страницыSEX DETERMINATION MECHANISMSManojkumar Ayyappan100% (1)
- Bikini Bottom Genetics Student WorksheetДокумент3 страницыBikini Bottom Genetics Student WorksheetRyan TelferОценок пока нет
- Practical 1 GeneticsДокумент20 страницPractical 1 GeneticsizzambrnОценок пока нет
- Week 3 Sci 8Документ45 страницWeek 3 Sci 8Joseph Gabriel Genuino CruzОценок пока нет
- Science9 q1 Mod3 SDOv2Документ32 страницыScience9 q1 Mod3 SDOv2Qwerty AnimeОценок пока нет
- Mendel's Laws of InheritanceДокумент37 страницMendel's Laws of InheritanceJudy RianoОценок пока нет
- Genetics and Evolution SUMMARY NOTESДокумент6 страницGenetics and Evolution SUMMARY NOTESPrabhОценок пока нет
- Chapter 18 Bio NotesДокумент14 страницChapter 18 Bio Notesgeezariana790Оценок пока нет
- Drosophila Lab ReportДокумент4 страницыDrosophila Lab ReportOdunayo KusoroОценок пока нет
- Science: Self - Learning ActivitiesДокумент22 страницыScience: Self - Learning ActivitiesYASER TOMASОценок пока нет
- 10th Week InheritanceДокумент9 страниц10th Week InheritanceJude Daniel AquinoОценок пока нет
- 9 Science Q1 W4Документ10 страниц9 Science Q1 W4Jewel ParedesОценок пока нет
- BMJ 40 13Документ8 страницBMJ 40 13Alvin JiwonoОценок пока нет
- Development Theories 1 2Документ66 страницDevelopment Theories 1 2Aaryan GuptaОценок пока нет
- BPH 25 and 26Документ9 страницBPH 25 and 26Sangram MohantyОценок пока нет
- Take Home Quiz COVID 2020Документ18 страницTake Home Quiz COVID 2020Jamie RuzickaОценок пока нет
- Mendelian Genetics PDFДокумент12 страницMendelian Genetics PDFjhaghjssОценок пока нет
- Practical Techniques PV92 PracticalДокумент5 страницPractical Techniques PV92 PracticalDavid OnarindeОценок пока нет