Академический Документы
Профессиональный Документы
Культура Документы
Kimball Et Al 2018, Figure 5
Загружено:
Abby KimballОригинальное название
Авторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Kimball Et Al 2018, Figure 5
Загружено:
Abby KimballАвторское право:
Доступные форматы
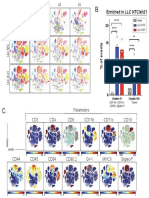
SPADE Settings (Cytobank)
A.
1. Define target number of nodes: 3. Select cellular population:
The automatic target number Select cellular population for
of nodes is 200, this can be analysis. Note: You cannot
user-defined select specific individual
samples or the number of
2. Select % of downsampling: events sampled
The automatic percent of 4. Select markers:
downsampled events is 10% Select all relevant
this can be user-defined phenotypic markers
for clustering
B. 200 Nodes C. 45 Nodes D. 45 Nodes E. 45 Nodes
(Representation #1) (Representation #2) (SPADE run #2)
5,217 cells
1 cell Cellular abundance
in each node is
expressed by
node size. 17,892 cells
22 cells
CD45
CD45 CD45
CD45 .97
1.41 1.41 17,892 cells
1.6
22 cells
In a SPADE tree, 6.59 6.59 19,447 cells 6.77
6.99
each cell is present 19 cells
in only a single node.
Marker intensity is The shape and orientation Sequential SPADE trees can look different
expressed by node color. of a SPADE tree is arbitrary between runs, in shape, node size,
and can be manually expression, and connections.
changed by the user.
F. # of markers used for clustering
Lineage Markers (10) All Markers (35)
Samples
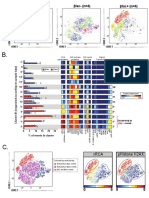
G.
X-shift Informed Target Nodes
IL10KO #4
IL10KO #3
IL10KO #2
IL10KO #1
B6 #4
B6 #3
B6 #2
B6 #5
B6 #1
CD45
# of target nodes
89 nodes 45 nodes
13,442.92 cells 17,892 cells
1 cell 22 cells
CD19
Automatic Target Nodes (200)
CD4
Parameters
CD8
5,217 cells
1 cell
6,509.84 cells
1 cell
CD11c
Multiple nodes containing only 1 cell
can indicate tree overfragmentation.
NKp46
H.
CD11b+, CD64+
What are the cellular CD64
CD4+ phenotypes of the
CD11b+, CD64-
various clusters?
CD45-
CD45+ CD11b
CD19+
NKp46+
Phenotypes are defined
CD8+ by marker intensity How consistent are
How do SPADE trees from
replicate samples in node
different experimental
size and expression?
groups differ in node Figure 5
size and expression?
Kimball et al
Вам также может понравиться
- LIGGGHTS Tutorial PDFДокумент18 страницLIGGGHTS Tutorial PDFTomas Dreves BazanОценок пока нет
- DNA Profiling: DNA Profiling (Also Called DNA Fingerprinting, DNA Testing, or DNA Typing) IsДокумент3 страницыDNA Profiling: DNA Profiling (Also Called DNA Fingerprinting, DNA Testing, or DNA Typing) IsShaik MoqtiyarОценок пока нет
- An Introduction To Computing With Neural NetsДокумент19 страницAn Introduction To Computing With Neural NetsAditya AgarwalОценок пока нет
- NewNode Analytics DocumentationДокумент38 страницNewNode Analytics DocumentationdangersamОценок пока нет
- Kimball Et Al 2018, Figure 1Документ1 страницаKimball Et Al 2018, Figure 1Abby KimballОценок пока нет
- Nallamothu URTC2021posterДокумент1 страницаNallamothu URTC2021posterSirihaasa NОценок пока нет
- Fibrilación Atrial, Dominio y Adaptación Acercamiento en Base A Una Red NeuronalДокумент6 страницFibrilación Atrial, Dominio y Adaptación Acercamiento en Base A Una Red NeuronalRafa LeónОценок пока нет
- ML DL AlgorithmДокумент11 страницML DL AlgorithmNguyen Quang HuyОценок пока нет
- CH 07Документ57 страницCH 07chandu mОценок пока нет
- SelexOn MeterManualДокумент36 страницSelexOn MeterManualquankddОценок пока нет
- Androvision®: Technical Data SheetДокумент9 страницAndrovision®: Technical Data SheetTulus MaulanaОценок пока нет
- Consolidate Complex Molecular Diagnostics Testing Needs in One Simple-to-Use and Flexible SolutionДокумент2 страницыConsolidate Complex Molecular Diagnostics Testing Needs in One Simple-to-Use and Flexible SolutionAli FirmanОценок пока нет
- Use of Deep Neural Networks in Automated DetectionДокумент29 страницUse of Deep Neural Networks in Automated DetectionARSHIA REHMAN100% (1)
- New Node Performance DashboardДокумент38 страницNew Node Performance DashboardJuan C LeonОценок пока нет
- An Introduction' To Computing With Neural Nets: Richard P. LippmannДокумент19 страницAn Introduction' To Computing With Neural Nets: Richard P. LippmannSathish Kumar KarneОценок пока нет
- HCS CEIVD FS GL-GN-2200013-r2Документ1 страницаHCS CEIVD FS GL-GN-2200013-r2WissamMamouОценок пока нет
- Serstech Indicator100 RAMANДокумент2 страницыSerstech Indicator100 RAMANabdurahman143Оценок пока нет
- Two-Stage Conditional Chest X-Ray Radiology Report GenerationДокумент6 страницTwo-Stage Conditional Chest X-Ray Radiology Report Generationns8c8ftpgjОценок пока нет
- Tube Inspection Probe Catalogue EN 201204 PDFДокумент52 страницыTube Inspection Probe Catalogue EN 201204 PDFIrawan EkklesiaОценок пока нет
- Technote Infinium Genotyping Data AnalysisДокумент10 страницTechnote Infinium Genotyping Data AnalysisANJU THOMASОценок пока нет
- App Data UserImages File Php2620 Lec12 ChIPseqДокумент24 страницыApp Data UserImages File Php2620 Lec12 ChIPseqJamesОценок пока нет
- 070 - Searching For Effective Neural Network Architectures For Heart Murmur Detection From PhonocardiogramДокумент4 страницы070 - Searching For Effective Neural Network Architectures For Heart Murmur Detection From PhonocardiogramDalana PasinduОценок пока нет
- DropoutДокумент14 страницDropoutAhmed EmadОценок пока нет
- Problem Bank 01: Assignment IДокумент9 страницProblem Bank 01: Assignment IUdayChanderAmbatiОценок пока нет
- Drawing Graphs With: NeatoДокумент11 страницDrawing Graphs With: NeatoJose Perez GarciaОценок пока нет
- Training Course - 5G RAN3.0 ANRДокумент40 страницTraining Course - 5G RAN3.0 ANRVVLОценок пока нет
- Optimum Architecture of Neural Networks Lane Following SystemДокумент6 страницOptimum Architecture of Neural Networks Lane Following SystemJ.Оценок пока нет
- Kimball Et Al 2019, Figure 2Документ1 страницаKimball Et Al 2019, Figure 2Abby KimballОценок пока нет
- Musical Gesture Recognition Using Machine Learning and Audio DescriptorsДокумент5 страницMusical Gesture Recognition Using Machine Learning and Audio DescriptorsNagendra NagshaОценок пока нет
- Soft Core Processor MicroarchitectureДокумент18 страницSoft Core Processor MicroarchitectureavinashdaraОценок пока нет
- CMAC Neural NetworksДокумент6 страницCMAC Neural NetworksijmremОценок пока нет
- Mini Project B20CS061Документ16 страницMini Project B20CS061THANMAI MITTAPELLYОценок пока нет
- Matlab Analysis of Eeg Signals For Diagnosis of Epileptic Seizures by Ashwani Singh 117BM0731 Under The Guidance of Prof. Bibhukalyan Prasad NayakДокумент29 страницMatlab Analysis of Eeg Signals For Diagnosis of Epileptic Seizures by Ashwani Singh 117BM0731 Under The Guidance of Prof. Bibhukalyan Prasad NayakAshwani SinghОценок пока нет
- Avenio Ctdna Analysis Kits Performance Across Illumina Sequencing PlatformsДокумент4 страницыAvenio Ctdna Analysis Kits Performance Across Illumina Sequencing PlatformspappuОценок пока нет
- Literature ReviewДокумент3 страницыLiterature Reviewrr0734Оценок пока нет
- Prediction of Strength of Fly Ash Concrete: Dept of Civil EngineeringДокумент22 страницыPrediction of Strength of Fly Ash Concrete: Dept of Civil EngineeringShinning GarimaОценок пока нет
- Yumi Ono, Yoshifumi Onishi, Takafumi Koshinaka, Soichiro Takata, and Osamu HoshuyamaДокумент5 страницYumi Ono, Yoshifumi Onishi, Takafumi Koshinaka, Soichiro Takata, and Osamu HoshuyamaJuan Camilo Gomez SotoОценок пока нет
- Data Minig Final ProjectДокумент16 страницData Minig Final ProjectAdmas MamoОценок пока нет
- Gender & Emotion Classification PresentДокумент31 страницаGender & Emotion Classification PresenttensuОценок пока нет
- WCDMA Neighboring Cell Support (WNCS)Документ21 страницаWCDMA Neighboring Cell Support (WNCS)mahmoudОценок пока нет
- Ashwani Singh 117BM0731Документ28 страницAshwani Singh 117BM0731Ashwani SinghОценок пока нет
- MFC in Emeraude Cheat SheetДокумент1 страницаMFC in Emeraude Cheat SheetYovaraj KarunakaranОценок пока нет
- Direction Finders 51: Introduction Into Theory of Direction FindingДокумент4 страницыDirection Finders 51: Introduction Into Theory of Direction FindingDimas RioОценок пока нет
- Vibration Analyzer, Data Collector & Dynamic BalancerДокумент8 страницVibration Analyzer, Data Collector & Dynamic Balancerwongpengchiong7205Оценок пока нет
- Bit-Wise Arithmetic Coding For Data CompressionДокумент16 страницBit-Wise Arithmetic Coding For Data CompressionperhackerОценок пока нет
- Vdocuments - MX Liggghts TutorialДокумент18 страницVdocuments - MX Liggghts Tutorialaashay professionalОценок пока нет
- DNA - STRIP Technology: Reliable Diagnostic Assays For Your ConvenienceДокумент3 страницыDNA - STRIP Technology: Reliable Diagnostic Assays For Your ConveniencejumaОценок пока нет
- Advanced Readout Integrated Circuit Signal ProcessingДокумент11 страницAdvanced Readout Integrated Circuit Signal ProcessingOmkar KatkarОценок пока нет
- EMR Final 2022Документ4 страницыEMR Final 2022foudaomar474Оценок пока нет
- Neural Observer Based Approach To FaultДокумент6 страницNeural Observer Based Approach To FaultHB RIMОценок пока нет
- Brochure Real-Time PCR ENДокумент4 страницыBrochure Real-Time PCR ENadonis pascualОценок пока нет
- Microarray Image Analysis and Gene Expression Ratio StatisticsДокумент42 страницыMicroarray Image Analysis and Gene Expression Ratio StatisticsSamir SabryОценок пока нет
- Efficient Solution Technique For Dendritic Channel Networks Using FEMДокумент10 страницEfficient Solution Technique For Dendritic Channel Networks Using FEMAMAN GUPTAОценок пока нет
- Uc Davis NMR Facility VNMRJ Short Manual For Varian/Agilent NMR SpectrometersДокумент45 страницUc Davis NMR Facility VNMRJ Short Manual For Varian/Agilent NMR SpectrometersEthan MatthewsОценок пока нет
- Mesh Analysis: Find The Branch Currents Using Mesh AnalysisДокумент4 страницыMesh Analysis: Find The Branch Currents Using Mesh AnalysisJiya AssijaОценок пока нет
- Mu-Fashion: Multi-Resolution Data Fusion Using Agent-Bearing Sensors in Hierarchically-Organized NetworksДокумент33 страницыMu-Fashion: Multi-Resolution Data Fusion Using Agent-Bearing Sensors in Hierarchically-Organized NetworkshaddanОценок пока нет
- Programming The M68000 1983 Addison-Wesley Publishing CompanyДокумент84 страницыProgramming The M68000 1983 Addison-Wesley Publishing CompanyAFK MasterОценок пока нет
- H.264 Analyzer - FinalPresentationДокумент30 страницH.264 Analyzer - FinalPresentationapi-19481404Оценок пока нет
- RnpregresskernelДокумент24 страницыRnpregresskernelDaniel Redel SaavedraОценок пока нет
- Nonnegative Matrix and Tensor Factorizations: Applications to Exploratory Multi-way Data Analysis and Blind Source SeparationОт EverandNonnegative Matrix and Tensor Factorizations: Applications to Exploratory Multi-way Data Analysis and Blind Source SeparationОценок пока нет
- Oko Et Al 2019, Figure 8Документ1 страницаOko Et Al 2019, Figure 8Abby KimballОценок пока нет
- Neuwelt Et Al, Figure 2Документ1 страницаNeuwelt Et Al, Figure 2Abby KimballОценок пока нет
- Kimball Et Al 2019, Figure 7Документ1 страницаKimball Et Al 2019, Figure 7Abby KimballОценок пока нет
- Kimball Et Al 2019, Figure 2Документ1 страницаKimball Et Al 2019, Figure 2Abby KimballОценок пока нет
- Kimball Et Al 2019, Figure 4Документ1 страницаKimball Et Al 2019, Figure 4Abby KimballОценок пока нет
- Kimball Et Al 2019, Figure 3Документ1 страницаKimball Et Al 2019, Figure 3Abby KimballОценок пока нет
- Kimball Et Al 2018, Figure 9Документ1 страницаKimball Et Al 2018, Figure 9Abby KimballОценок пока нет
- Kimball Et Al 2019, Figure 1Документ1 страницаKimball Et Al 2019, Figure 1Abby KimballОценок пока нет
- Kimball Et Al 2018, Figure 7Документ1 страницаKimball Et Al 2018, Figure 7Abby KimballОценок пока нет
- Kimball Et Al 2018, Figure 8Документ1 страницаKimball Et Al 2018, Figure 8Abby KimballОценок пока нет
- Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectДокумент1 страницаGraph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectAbby KimballОценок пока нет
- Bullock Et Al 2018, Figure 6Документ1 страницаBullock Et Al 2018, Figure 6Abby KimballОценок пока нет
- Bullock Et Al 2018, Supplemental Figure 5Документ1 страницаBullock Et Al 2018, Supplemental Figure 5Abby KimballОценок пока нет
- Kimball Et Al 2018, Figure 2Документ1 страницаKimball Et Al 2018, Figure 2Abby KimballОценок пока нет
- Berger Et Al 2019, Figure 2Документ1 страницаBerger Et Al 2019, Figure 2Abby KimballОценок пока нет
- 2012 Dse Bio 1 MS 1Документ6 страниц2012 Dse Bio 1 MS 1peakОценок пока нет
- EcoRI - WikipediaДокумент12 страницEcoRI - WikipediaArjun ArulvelОценок пока нет
- Lymphocyte TraffickingДокумент8 страницLymphocyte TraffickingAbhishek KumarОценок пока нет
- The Powers of The MindДокумент1 страницаThe Powers of The MindKen Walker100% (1)
- 22 23 Filipino 10 TOS 4th GRADINGДокумент4 страницы22 23 Filipino 10 TOS 4th GRADINGAbegail ReyesОценок пока нет
- Review Questions For Online Pre Final ExamДокумент5 страницReview Questions For Online Pre Final ExamReygie MataОценок пока нет
- Crop Protection 1 Syllabus in The New NormalДокумент14 страницCrop Protection 1 Syllabus in The New NormalJESRYL PAULITE100% (3)
- ROSELA, ERLAINE MARIE Blood-LabДокумент4 страницыROSELA, ERLAINE MARIE Blood-LabPadoОценок пока нет
- Scientific Paper MicrosДокумент7 страницScientific Paper MicrosThe Gaile-squared ShopОценок пока нет
- Bio2 Test 3Документ16 страницBio2 Test 3Carrie NortonОценок пока нет
- 10 Biology Structural Organization of AnimalsДокумент2 страницы10 Biology Structural Organization of AnimalsHasan shaikhОценок пока нет
- EMTEA Study Guide FinalДокумент4 страницыEMTEA Study Guide FinalJuan GironaОценок пока нет
- Term Paper ON: Transgenic PlantsДокумент17 страницTerm Paper ON: Transgenic PlantsVikal RajputОценок пока нет
- 60 Herbs Against CancerДокумент33 страницы60 Herbs Against CancerAnonymous iPygec100% (1)
- Form 2 Science Chapter 1 NoteДокумент4 страницыForm 2 Science Chapter 1 Noteseek_4_happinesss100% (1)
- GlioblastomaДокумент452 страницыGlioblastomaDesmondОценок пока нет
- Human Physiology The Basic of MedicineДокумент1 816 страницHuman Physiology The Basic of Medicinebranislava_vuković100% (3)
- Cactus (Opuntia Ficus-Indica) : A Review On Its Antioxidants Properties and Potential Pharmacological Use in Chronic DiseasesДокумент9 страницCactus (Opuntia Ficus-Indica) : A Review On Its Antioxidants Properties and Potential Pharmacological Use in Chronic DiseasesAidОценок пока нет
- DNA by Dr. Ajit Kulkarni-59923560Документ16 страницDNA by Dr. Ajit Kulkarni-59923560Priyanka MovaliyaОценок пока нет
- Pre Biology POSN - Examination PLUSДокумент108 страницPre Biology POSN - Examination PLUSTetee TetatОценок пока нет
- Microbiology Exam 3Документ9 страницMicrobiology Exam 3Sam SchlueterОценок пока нет
- Iji2013 151028Документ8 страницIji2013 151028Kershaun MathewОценок пока нет
- Assurance Gds Brochure Br2694en MKДокумент4 страницыAssurance Gds Brochure Br2694en MKFatimaezzahraa BouaalamОценок пока нет
- SPECIMEN COLLECTION and Processing For BACTERIOLOGY PDFДокумент19 страницSPECIMEN COLLECTION and Processing For BACTERIOLOGY PDFLPDF010596Оценок пока нет
- CATS & DOGS Nutrition PDFДокумент71 страницаCATS & DOGS Nutrition PDFErald Renz LaurenteОценок пока нет
- Kami Export - Mya Bennett - Intro To Cells Recap by Amoeba SistersДокумент2 страницыKami Export - Mya Bennett - Intro To Cells Recap by Amoeba SistersMya BennettОценок пока нет
- Dsa FLXM Pi en 00Документ7 страницDsa FLXM Pi en 00yantuОценок пока нет
- Flowers Chap17Документ15 страницFlowers Chap17Cheret LengОценок пока нет
- Cell Biology Lec No 6 ChloroplastДокумент18 страницCell Biology Lec No 6 ChloroplastsaleemОценок пока нет