Академический Документы
Профессиональный Документы
Культура Документы
Molecular Biology: Lesson 3: Dna Repair and Recombination
Загружено:
jeffreyОригинальное название
Авторское право
Доступные форматы
Поделиться этим документом
Поделиться или встроить документ
Этот документ был вам полезен?
Это неприемлемый материал?
Пожаловаться на этот документАвторское право:
Доступные форматы
Molecular Biology: Lesson 3: Dna Repair and Recombination
Загружено:
jeffreyАвторское право:
Доступные форматы
S
E
MOLECULAR BIOLOGY 2 nd M
LESSON 3: DNA REPAIR AND RECOMBINATION
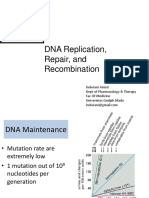
DNA POLYMERASE PROOFREADING A. SPONTANEOUS DNA DAMAGE
• Oxidative Damage
• Hydrolytic Attack
• Methylation
• This figure shows the representation of the site where
the DNA damage occur.
• Red arrow indicates Oxidative damage, Blue arrow
• DNA polymerase has 2 types: indicates Hydrolytic attack and the Green arrow
o The polymerizing site indicates the Methylation.
▪ Adds new nucleotide
o The Editing site METHYLATION
▪ Detects bases which are mispaired. If it
detects the mispaired, the DNA polymerase
has an exonuclease activity to remove the
incorrect nucleotide.
• Basically, the DNA polymerase has an own
proofreading mechanism and it allows us to have
low mutation rate.
o Eukaryotic DNA polymerases make errors every
10,000-100,000 nucleotides
o 1 mutation per 10 billion nucleotide base pairs per • In methylation, due to harmful chemicals, methyl
cell generation groups are added to the nucleotide bases.
DNA DAMAGE AND REPAIR B. DEPURINATION
• The human cell loses about 18,000 purine bases
every day because of Depurination.
• This image shows how DNA repair happened. • Depurination is where your glycosyl linkage of
purine to deoxyribose hydrolyzed
• Researchers from Harvard school of Medicine
labeled the end of the DNA with Fluorescent marker
o In depurination, it removes the purine. The
purines are Guanine and Adenine.
and they observed that once that the DNA ends
reconnect, they changed in color.
o Depurination happens only in Purines, Guanine
and Adenine.
• Without DNA repair, spontaneous DNA damage
would rapidly change the DNA sequences even on • The Glycosyl linkage (links to sugar and nucleotide
their normal cell condition the DNA is susceptible to bases) is hydrolyzed. The water acts upon glycosyl
damage. linkage therefore displacing the nucleotide base.
• These damages may lead to mutation and if they are o When the glycosyl linkage hydrolyze, the purine
not repaired, they will cause certain diseases. will be removed.
• The purines are substituted with hydroxyl group. o If you deaminate the Adenine, you will have
“Hypoxanthine”
• In Guanine, the amino group is hydrolyzed therefore
displacing the amino group and will change to
“Xanthine”
o If you deaminate the Adenine, you will have
“Xanthine”
• In depurination, once it is happened, there is mutation
specifically the lesion
• When the nucleotide Is depurinated, the purine • In Pyrimidines, there is Cytosine. The amino group
(Adenine) is removed. will hydrolyze and will be removed. It will form Uracil
• During the DNA replication, when it uses the template • In Thymine, there is no deamination because there
with mutation, it will have a mutation that will occur. is no amino group.
The new DNA will no longer have A-T nucleotide
because of depurinate nucleotide.
C. DEAMINATION
• Deamination is the conversion of nucleotide to
unnatural products by hydrolysis of the amino group.
o In deamination, the amino group is hydrolyzed,
one it is hydrolyzed, it is removed. So, it will
change Cytosine to Uracil. • Deamination creates mutation
• The most common example is Cytosine o Deamination = Substitution Mutation
o Depurination = Deletion Mutation
o In deamination, once the Cytosine is deaminated
to Uracil, there is a change in the template
strand. When it uses the template strand for DNA
synthesis, there will have a new nucleotide base.
D. PYRIMIDINE DIMERS
• In Pyrimidine dimers, the UV light affects the
• The Adenine has amino group and the amino group
nucleotide bases. They formed a covalent bond and
is hydrolyzed therefore displacing the amino group
and you will have unnatural products called this covalent bond, once it is formed, it becomes a
Dimer.
“Hypoxanthine”
• The most common dimer is the Thymine dimer. This
thymine dimer forms a kink in the DNA strand.
E. BREAKS IN THE DNA
• Breaks in the DNA is caused by X-rays or Gamma-
rays or other environmental factor.
• Single strand break and Double strand break are
the damages in the DNA.
IF THE DNA DAMAGES ARE NOT REPAIRED
AND ACCUMULATE, DISEASE WILL OCCUR
• If the Thymine dimer is still carried on in the
transcription, it can cause diseases. That’s why we
need to repair the damages. Luckily, our cells have
the ability to repair DNA damages.
• Ex. Xeroderma pigmentosum
DNA DOUBLE HELIX IS READILY
REPAIRED
• In this figure, it detects that there is a damage,
o DNA polymerase will be released and will
changed to translesion DNA polymerase
o This translesion DNA polymerase is in the form of
DNA polymerase eta in eukaryotes
o What it does, it continues to replicate the DNA
o The translesion DNA polymerase has the ability
to put nucleotide sequences in the growing
strand even though it has thymine dimer in the
template strand.
▪ Thymine dimers is the usually DNA damage
▪ Add correct base pair in the form of Adenine
• DNA damages are easily repair because the DNA
double helix is very ideal. REPAIR OF SINGLE STRAND BREAKS
o The DNA double helix is a complementary • Mismatch Repair
strand, so you can use either of the strand to o The back-up repair during DNA replication
correct the mutated strand. o genetic defects (Lynch Syndrome)
• Base Excision Repair
TRANSLESION POLYMERASE o carcinogenic exposure (such as tobacco smoking
• The replicative polymerase is switched out for and/or diesel exhaust)
specialized DNA polymerases and replicates past the • Nucleotide Excision Repair
damage. o Most common used repair for DNA damages
• DNA polymerase n (eta) which are bulky
o DNA polymerase used in eukaryotes ▪ For Thymine dimers
o UV light (Xeroderma pigmentosum)
A. MISMATCH REPAIR B. BASE EXCISION REPAIR
• In this figure, A mismatched is detected in the newly
synthesized DNA
o Thymine is the complementary base pair but the • Deamination converts a cytosine base into Uracil
added nucleotide strand is guanine • The Uracil is detected and removed, leaving a base-
• The new DNA strand is cut and the mispaired less nucleotide
nucleotide and its neighbor are removed. • The Sugar phosphate backbone is also removed.
• The missing patch is replaced with correct When the base-less nucleotide is removed, leaving a
nucleotides by a DNA polymerase. small gap in the DNA
o Once the section is removed, the DNA • The hole is filled with the right base by a DNA
polymerase puts the right nucleotide bases polymerase, and the gap is sealed by a ligase.
• A DNA ligase seals the gap in the DNA backbone.
• In mismatch repair, it has a protein called MutS and
MutL complex. It detects the mismatch in the DNA
• The MutH detects certain sequence in the DNA called
GATC. If the GATC is detected, the certain sequence
will bind to the complex and it bends the DNA.
o When It is bended, the MutH makes a cut unto
the mismatch base pair • DNA glycosylases
o Once it makes a cut the DNA polymerase can o “flipping out”
now add the nucleotide base pair which are ▪ It flips out the nucleotide base to go outside
correct. o recognizes and removes the unnatural nucleotide
o An enzyme that detects the wrong in the template
• AP endonuclease
o “missing tooth”
▪ Removes phosphodiester and sugar
phosphate that makes it to look like a missing
tooth.
o Removes the sugar-phosphate backbone • Excision nuclease
▪ It produces a gap in the DNA o Detects the error
▪ The gap is filled with new nucleotides and the o It marks the section of the DNA to be remove
nick is sealed by DNA ligase • DNA Helicase
o Removes the strand of the DNA which has the
C. NUCELOTIDE EXCISION REPAIR pyrimidine dimers
• “Bulky lesions”
• DNA Polymerase
o covalent reactions of DNA bases with large
o Adds the correct DNA bases
hydrocarbons (e.g. carcinogen benzopyrene
found in tobacco smoke, coal tar, and diesel • DNA Ligase
exhaust) o Corrects the gap between the strands
o pyrimidine dimers caused by sunlight
REPAIR OF DOUBLE STRAND BREAK
• Non-homologous End Joining
• Homologous Recombination
A. NON-HOMOLOGOUS END JOIN
• In the figure, UV radiation produces a thymine dimer
• Once the dimer has been detected, the surrounding • Scientists in Harvard school of Medicine uses the
DNA is opened to form a bubble technique Fluorescence Resonance Energy
• Enzymes cut the damaged region out of the bubble Transfer (FRET) microscopy, that once the DNA
• DNA polymerase replaces the excised (cut-out) DNA connects it changes the color to Red.
with new correct nucleotide sequence and a ligase o In this way, we can observe the in between
seals the backbone phases of the non-homologous joining.
• Non-homologous end joining is very simple
because it just glues the DNA strand back together,
usually with a small mutation at the break site.
• It is also called “Quick end dirty” because it just
• Enzymes responsible for Nucleotide excision repair: connects the end of the DNA
B. HOMOLOGOUS RECOMBINATION
• Once the strand exchange occurs, the repair
polymerase, synthesizes the DNA (represented in
• In homologous recombination, the damaged region is green) using the undamaged DNA as template
replaced via recombination, using sequences copied
• Invading strand release and broken helix re-formed.
from the homologue.
• DNA synthesis continues using strands from
damaged DNA as template
• And lastly, it will have DNA Ligation
• Based in the figure, it has a double strand break and
when it makes a daughter DNA, it will produce 2
daughter DNA double helix.
o The 1st daughter DNA has a double strand break
o The nuclease will digest the region of the
daughter DNA that has double strand break and
it will have a strand exchange from the other
daughter DNA will go inside to copy and replaced • Repair of broken replication fork
the damaged DNA. o The nuclease will degrade the part which has a
• Homologous recombination nick or damage and once the nuclease degrade,
o General term for crossing over of DNA section to It will have an overhang which goes to the strand
another DNA section (alleles with same which is damage. And it is termed as “Grand
hereditary information) in Myosis. Exchange”
o Genetic information from parents will combine ▪ In Grand exchange, the damage DNA uses
o In this case, Homologous recombination is used the undamaged DNA as a template
to repair DNA with double strand break o Once the strand breakage and additional
synthesis happen, It can already have a DNA
replication.
o You can see at the last part of the picture that the
Replication fork resumes/continues to work.
CASE STUDY • In Normal tissue (blue line) microsatellite markers
• Hereditary Non-polyposis Colon Cancer do not match to the Tumor tissue (red); therefore, it
has Instability.
• Xeroderma Pigmentosum
• The MSS (Stable) matches the microsatellite
• BRCA1 and BRCA2
HEREDITARY NON-POLYPOSIS COLON
CANCER (HNPCC)
• Lynch Syndrome
o Genetic disorder that causes an increased risk of
developing cancer
o Autosomal dominant
o Mutation in the genes involved in DNA repair
▪ Mismatch Repair Genes (MMR genes)
• Can be diagnosed using Microsatellite Instability
Testing (MSI)
o The presence of MSI is an evidence of mismatch
repair gene is not functioning properly
• Microsatellite
o A condition in which it has hyper mutability in
impaired DNA. XERODERMA PIGMENTOSUM
o A structure which repeated nucleotide mostly • Children of the moon
often seen as GTCA. • It is a rare hereditary disease which children who
o Are repeats in DNA and normal cells. inherited the disease is very sensitive to the sun
o When the mismatch repair is working properly, ultra-violet light / radiation.
the microsatellite will all properly replicates. o Exposure even a few minutes in the sunlight is
o Researchers found out that when we look at the enough to cause severe burning in the skin or it
microsatellite, we can actually determine if the caan cause skin cancer.
person has an HNPCC. • Their suit is designed by NASA to prevent them from
the UV radiation.
• In the Normal satellite replication, you will see the
right replication of the DNA
• In the Abnormal mismatch repair defect, the
microsatellite has a variability in sizes
o Variability in microsatellite is called
Microsatellite Instability.
MICROSATELLITE INSTABILITY TESTING
BY PCR
• In Microsatellite Instability Testing, it amplifies the
Microsatellite (repeats). • UV radiation can cause DNA damage by triggering
• In MSI-H (Height), it is the samples with microsatellite pyrimidine dimer formation
instability, therefore the variability of the microsatellite • Excision repair to remove dimer formation is
is high. dysfunctional
• The Fam, Hex and Texas red are Chromophores • Mutations in seven genes, XPA to XPG, responsible
that use in PCR. for the excision repair pathway
o These chromophores are the one who gives o Their body has no capability to exhibit the
signal if it detects the section of DNA excision repair, that’s why the dimers accumulate
• The Bat 25, NR24, NR21, NR27, and Bat25 are only to cause a cancer.
a microsatellite marker. These are the ones amplified.
• Another type of Xeroderma Pigmentosum is the BREAST CANCER
Mutations in the eighth gene called XPV produce
an alternate form of xeroderma pigmentosum
• Affects DNA polymerase ɳ (eta) responsible for
translesion synthesis
• 2 DNA repair mechanisms affected in XP:
1. Excision Repair
2. Translesion Synthesis
• For diagnosis of breast cancer, the ff can be used:
o Breast Exam
o Needle Biopsy
o MRI
SUMMARY
• Discussed about the different types of DNA damage.
• Learned the various ways in which DNA repair is
conducted for all kinds of damage, such as dimers,
• Xeroderma Pigmentosum can be diagnosed by single and double strand DNA breaks.
o Skin Biopsy • Discussed the mechanism behind DNA
o Genetic Testing recombination.
▪ By giving a sample of the DNA to see if it has • Studied diseases relating to the dysfunction of DNA
a mutation in the genes involve in the repair.
xeroderma pigmentosum
• Very rare, diagnosis not yet fully developed. Usually,
this disease is discovered in grown-ups only.
BRCA1 AND BRCA2
• Brca1 and Brca2 gene compromise the homologous
recombination repair and causes Hereditary Breast
and Ovarian Cancer
o Mutations in Brca1 and Brca2 results to ovarian
and breast cancer because of damage
accumulation
• Brca1 and Brca2 aids in the homologous
recombination.
• Brca1 is the protein that’s initiate or calls the other
enzyme to repair the DNA damage
• Brca2 holds a protein mRad51 which facilitates to
enter the DNA strand to the homologous repair
Вам также может понравиться
- A Level Biology For OCR A (Double Paged)Документ360 страницA Level Biology For OCR A (Double Paged)squishyboitae33% (3)
- Amino Acid MetabolismДокумент46 страницAmino Acid Metabolismrsmbgss100% (1)
- Parasitology TablesДокумент9 страницParasitology Tables2013SecB92% (25)
- Epidemiology & of Cancer in India: EtiologyДокумент20 страницEpidemiology & of Cancer in India: Etiologysuman singhОценок пока нет
- The HallMarks of CancerДокумент33 страницыThe HallMarks of CancerAmaan Khan100% (1)
- Cancer and CarcinosinДокумент12 страницCancer and CarcinosinDr. Nur-E-Alam RaselОценок пока нет
- Viruses and RetrovirusesДокумент10 страницViruses and RetrovirusesJose Manuel MartinezОценок пока нет
- Ga Matlab ToolДокумент3 страницыGa Matlab Toolकुँवर रुपेन्र्द प्रताप सिंह हाड़ाОценок пока нет
- The Prime Cause and Prevention of Disease WarburgДокумент3 страницыThe Prime Cause and Prevention of Disease WarburgMichael DietrickОценок пока нет
- Dna Mutations Practice WorksheetДокумент3 страницыDna Mutations Practice Worksheetapi-32230392250% (2)
- Final Exam On Biostatistics and Epidemiology: Test IДокумент1 страницаFinal Exam On Biostatistics and Epidemiology: Test IjeffreyОценок пока нет
- Appendix 45 - Itinerary of TravelДокумент4 страницыAppendix 45 - Itinerary of TravelAngeli Lou Joven Villanueva100% (2)
- DNA Repair Mechanisms-1 PDFДокумент21 страницаDNA Repair Mechanisms-1 PDFMehak preetОценок пока нет
- NEET Botany Ecosystem Questions SolvedДокумент18 страницNEET Botany Ecosystem Questions SolvedYogesh AhireОценок пока нет
- Dnareplication 151218084840Документ145 страницDnareplication 151218084840RIYA MUNJALОценок пока нет
- DNA Damage and Repair MechanismsДокумент62 страницыDNA Damage and Repair MechanismsnikkiОценок пока нет
- Clinical Chemistry Reviewer by MTRДокумент56 страницClinical Chemistry Reviewer by MTRRyan Pan95% (19)
- Boron RomanaДокумент1 284 страницыBoron RomanaDaniel BaciuОценок пока нет
- DLP 1 For 3rd GradingДокумент3 страницыDLP 1 For 3rd GradingMarivic Guillermo - LongaresОценок пока нет
- Methylene Blue CAS No 61-73-4: Material Safety Data Sheet Sds/MsdsДокумент7 страницMethylene Blue CAS No 61-73-4: Material Safety Data Sheet Sds/MsdsjeffreyОценок пока нет
- Mechanism of Enzyme CatalysisДокумент18 страницMechanism of Enzyme CatalysisBuhlebenkosi Jackie BhebheОценок пока нет
- Vitamin C - A Stem Cell Promoter in Cancer Metastasis and ImmunotherapyДокумент7 страницVitamin C - A Stem Cell Promoter in Cancer Metastasis and Immunotherapykumar23Оценок пока нет
- Current Concepts in ElectrotherapyДокумент8 страницCurrent Concepts in ElectrotherapyAndri AajОценок пока нет
- Cleavable LinkersДокумент12 страницCleavable LinkersSrinivasa Reddy Telukutla100% (1)
- 5G Wireless TechnologyДокумент13 страниц5G Wireless TechnologyHani TaHaОценок пока нет
- 1.1 Enzymology (Bravo)Документ11 страниц1.1 Enzymology (Bravo)Arman Carl DulayОценок пока нет
- 2011 Urine Therapy J of NephrolДокумент3 страницы2011 Urine Therapy J of NephrolCentaur ArcherОценок пока нет
- ChemPhysics MCAT NotesДокумент14 страницChemPhysics MCAT NotesChris HuebnerОценок пока нет
- Electrochemistry Lect Notes CambridgeДокумент4 страницыElectrochemistry Lect Notes Cambridgeavatar_75Оценок пока нет
- The Chemistry of The Thiol Group (Part 2) (PDFDrive) PDFДокумент246 страницThe Chemistry of The Thiol Group (Part 2) (PDFDrive) PDFAmel AyadiОценок пока нет
- Cell MembraneДокумент24 страницыCell Membraneapi-3733156Оценок пока нет
- Caffeine & Creativity 1 PDFДокумент8 страницCaffeine & Creativity 1 PDFTai Kah KiangОценок пока нет
- CSIM2.24 - Signal TransductionДокумент6 страницCSIM2.24 - Signal TransductionAinahMahaniОценок пока нет
- Using Technology To Study Cellular and Molecular BiologyДокумент138 страницUsing Technology To Study Cellular and Molecular BiologybheeshmatОценок пока нет
- DNA Damage and RepairДокумент15 страницDNA Damage and RepairYeasmin AkhtarОценок пока нет
- Lecture 5 DNA Mutation and DNA Repair-2Документ65 страницLecture 5 DNA Mutation and DNA Repair-2Constance WongОценок пока нет
- Isolation and Characterization of DNAДокумент5 страницIsolation and Characterization of DNAgeorgiana0% (1)
- Perbaikan DnaДокумент33 страницыPerbaikan DnavictoryОценок пока нет
- Adv MBДокумент23 страницыAdv MBsreenidhi vОценок пока нет
- 1 Los Genes Son ADNДокумент24 страницы1 Los Genes Son ADNEmanuel Rodriguez ArrazateОценок пока нет
- Perbaikan Dna Mku 2018Документ40 страницPerbaikan Dna Mku 2018Ruman KhoirulwaroОценок пока нет
- DNA Damage Repair MechanismsДокумент40 страницDNA Damage Repair MechanismspriannaОценок пока нет
- Clase 9 DNA Reparation and Recombination-2Документ31 страницаClase 9 DNA Reparation and Recombination-2MacaCastilloОценок пока нет
- DNA Denaturation: Biochemistry Department. Course Code:701 Instructor: Dr/OmneyaДокумент16 страницDNA Denaturation: Biochemistry Department. Course Code:701 Instructor: Dr/OmneyaahmedalynasrОценок пока нет
- DNA Repair: Vipin ShankarДокумент32 страницыDNA Repair: Vipin ShankarMaximilian Magulye100% (1)
- DNA RepairДокумент36 страницDNA RepairMeenal MeshramОценок пока нет
- Isolation and Characterization of Nucleic Acids: Wheat BarleyДокумент3 страницыIsolation and Characterization of Nucleic Acids: Wheat BarleyAdrienne Nicole MerinaОценок пока нет
- Genetic Material-DNA: 6 November 2003 Reading:The Cell Chapter 5, Pages: 192-201Документ38 страницGenetic Material-DNA: 6 November 2003 Reading:The Cell Chapter 5, Pages: 192-201Laura VgОценок пока нет
- Extraction and Identification of DNAДокумент13 страницExtraction and Identification of DNAananyapandey6582Оценок пока нет
- Dna StructureДокумент30 страницDna StructurekaverinmohiteОценок пока нет
- MBY 364 NOTES - MergedДокумент22 страницыMBY 364 NOTES - Mergedroelienvv7Оценок пока нет
- DNA Replication, Repair, and RecombinationДокумент114 страницDNA Replication, Repair, and Recombinationviola septinaОценок пока нет
- Perbaikan Dna Mku 2021Документ50 страницPerbaikan Dna Mku 2021fita triastuti /ananda syifa ahmad syamilОценок пока нет
- DNA RNA Protein SynthesisДокумент41 страницаDNA RNA Protein Synthesisjohairah merphaОценок пока нет
- Nucleic Acid and Amio Acid Structure and FunctionДокумент47 страницNucleic Acid and Amio Acid Structure and FunctionMwanja MosesОценок пока нет
- Experiment 8 and 9 PDFДокумент17 страницExperiment 8 and 9 PDFKrizzi Dizon GarciaОценок пока нет
- Lecture 2 - DNA & ChromosomeДокумент74 страницыLecture 2 - DNA & ChromosomeConstance WongОценок пока нет
- DNA Repair Farina PDFДокумент38 страницDNA Repair Farina PDFSbahat FatimaОценок пока нет
- 3rd Sem Seminar - DNA Damage & RepairДокумент37 страниц3rd Sem Seminar - DNA Damage & RepairshararohitОценок пока нет
- DNA Damage & RepairДокумент48 страницDNA Damage & Repairbukan siapa siapaОценок пока нет
- 9 Photoactivated Dna Repair and Vit DДокумент34 страницы9 Photoactivated Dna Repair and Vit DMuhamad Ilfan AlwirudinОценок пока нет
- L20 - DNA Repair & RecombinationДокумент41 страницаL20 - DNA Repair & RecombinationLeroy ChengОценок пока нет
- DNA Denaturation: Biochemistry Department. Course Code:701 Instructor: Dr/OmneyaДокумент16 страницDNA Denaturation: Biochemistry Department. Course Code:701 Instructor: Dr/OmneyaAshik AdnanОценок пока нет
- Virtual Chemistry Lab DNA ExtractionДокумент4 страницыVirtual Chemistry Lab DNA ExtractionPedrosa NardОценок пока нет
- Dna Damage & RepairДокумент12 страницDna Damage & RepairDr Abhinav Manish MDОценок пока нет
- Molecular GeneticsДокумент2 страницыMolecular GeneticsFaith YeОценок пока нет
- Mb17 Dna RepairДокумент19 страницMb17 Dna Repairfaezeh zare karizakОценок пока нет
- Dna Replication - Prachi.sДокумент10 страницDna Replication - Prachi.sprachishnd74Оценок пока нет
- Molecular-Biology NotesДокумент2 страницыMolecular-Biology NotesAncestral garmentsОценок пока нет
- DNA Damage and RepairДокумент23 страницыDNA Damage and RepairbadrhashmiОценок пока нет
- Usi Sultan Activity6 CCДокумент3 страницыUsi Sultan Activity6 CCjeffreyОценок пока нет
- Ra 6425 Dangerous Drugs ActДокумент59 страницRa 6425 Dangerous Drugs ActjeffreyОценок пока нет
- Parasitology - Finals LABДокумент23 страницыParasitology - Finals LABjeffreyОценок пока нет
- Gram Positive Cocci: (+) Catalase (-)Документ4 страницыGram Positive Cocci: (+) Catalase (-)jeffreyОценок пока нет
- Intern Assessment Activity:: Medical Technology Internship Program - ChemistryДокумент3 страницыIntern Assessment Activity:: Medical Technology Internship Program - ChemistryjeffreyОценок пока нет
- Hormone Assays: Immuno-Serology SectionДокумент52 страницыHormone Assays: Immuno-Serology SectionjeffreyОценок пока нет
- Orange Best Workplace Safety Tips InfographicДокумент1 страницаOrange Best Workplace Safety Tips InfographicjeffreyОценок пока нет
- Identifying Artists of Different Periods and Their ArtworksДокумент12 страницIdentifying Artists of Different Periods and Their ArtworksjeffreyОценок пока нет
- Immunology and Serology Lecture 3 - Innate ImmunityДокумент10 страницImmunology and Serology Lecture 3 - Innate ImmunityjeffreyОценок пока нет
- Certificate of Employment For EmployeesДокумент5 страницCertificate of Employment For EmployeesjeffreyОценок пока нет
- Analysis of Urine and Other Body Fluids - , RMT Sputum & Bronchoalveolar Lavage (Bal)Документ11 страницAnalysis of Urine and Other Body Fluids - , RMT Sputum & Bronchoalveolar Lavage (Bal)jeffreyОценок пока нет
- Abarca, Jeffrey A. BSMT L1 (Bacteriology)Документ1 страницаAbarca, Jeffrey A. BSMT L1 (Bacteriology)jeffreyОценок пока нет
- Chapter 1 5 Super FinalOther ResearchДокумент176 страницChapter 1 5 Super FinalOther ResearchjeffreyОценок пока нет
- MT Laws Blood BankingДокумент59 страницMT Laws Blood BankingjeffreyОценок пока нет
- Module 1 EvaluationДокумент1 страницаModule 1 EvaluationjeffreyОценок пока нет
- Business Plan (Jeffrey A. Abarca BSMT H1)Документ63 страницыBusiness Plan (Jeffrey A. Abarca BSMT H1)jeffreyОценок пока нет
- Airborne PrecautionsДокумент13 страницAirborne PrecautionsjeffreyОценок пока нет
- Filipino Psychology (Panti Sisters) AnalysisДокумент2 страницыFilipino Psychology (Panti Sisters) AnalysisjeffreyОценок пока нет
- Mideo Cruz's PoliteismoДокумент3 страницыMideo Cruz's PoliteismojeffreyОценок пока нет
- Botong (Barringtonia Asiatica) Seed Extract As An Anti-Fungal Agent For Powdery Mildew in Eggplant (Solanum Melongena)Документ49 страницBotong (Barringtonia Asiatica) Seed Extract As An Anti-Fungal Agent For Powdery Mildew in Eggplant (Solanum Melongena)jeffreyОценок пока нет
- 108 PDFДокумент7 страниц108 PDFpuspa sariОценок пока нет
- Scaling Test - JawaharДокумент12 страницScaling Test - JawaharJawahar JawaharОценок пока нет
- Li2017 DisignДокумент14 страницLi2017 DisignDanyal2222Оценок пока нет
- Student Exploration: Digestive SystemДокумент4 страницыStudent Exploration: Digestive SystemNO POGGEGRОценок пока нет
- Latihan MitosisДокумент7 страницLatihan MitosisChe Mahani HussainОценок пока нет
- Organisation of Living Things Study PresentationДокумент26 страницOrganisation of Living Things Study PresentationRUIZ CANO EnriqueОценок пока нет
- Norditropin PresentationДокумент5 страницNorditropin PresentationCaralee Nelson HarpsøeОценок пока нет
- CHAPTER 3 Cellular Energy Transformation v1Документ70 страницCHAPTER 3 Cellular Energy Transformation v1Dhanrence AmuraoОценок пока нет
- Plant Transport Multiple Choice Qs PDFДокумент7 страницPlant Transport Multiple Choice Qs PDFMathula MuhundanОценок пока нет
- HydraДокумент3 страницыHydraMagdaleneОценок пока нет
- Leaky Blood Vessels - An Unknown Danger of COVID-19 Vaccination - Doctors For COVID EthicsДокумент2 страницыLeaky Blood Vessels - An Unknown Danger of COVID-19 Vaccination - Doctors For COVID EthicsHansley Templeton CookОценок пока нет
- Poe 6-176Документ1 страницаPoe 6-176suhani.gnupadhyayОценок пока нет
- NCERT Solution: Plant KingdomДокумент7 страницNCERT Solution: Plant KingdomJonathan Tolentino MercadoОценок пока нет
- 2-Autoimmune DiseasesДокумент29 страниц2-Autoimmune DiseasessoniaОценок пока нет
- Eukaryotic Gene Expression PDFДокумент2 страницыEukaryotic Gene Expression PDFDestinyОценок пока нет
- Anthropology Is A Science of HumankindДокумент2 страницыAnthropology Is A Science of HumankindSuman BaralОценок пока нет
- GEd 109 BiodiversityДокумент30 страницGEd 109 BiodiversityArisОценок пока нет
- Applications of Bionanotechnology in Nanomedicine: A Theoretical AnalysisДокумент6 страницApplications of Bionanotechnology in Nanomedicine: A Theoretical AnalysisPhilip KpaeОценок пока нет
- PiXL Knowledge Test QUESTIONS - AQA B1 CORE Science - Legacy (2016 and 2017)Документ6 страницPiXL Knowledge Test QUESTIONS - AQA B1 CORE Science - Legacy (2016 and 2017)Mrs S BakerОценок пока нет
- Concepts of BiofertilizersДокумент8 страницConcepts of BiofertilizersVasu MathuraОценок пока нет
- 8Документ3 страницы8api-412107487Оценок пока нет
- 03 Laporan Genetika IMITASI RATIO FENOTIPEДокумент8 страниц03 Laporan Genetika IMITASI RATIO FENOTIPEhananОценок пока нет
- Parts of Prokaryotic & Eukaryotic CellsДокумент7 страницParts of Prokaryotic & Eukaryotic CellsRose Yan JiОценок пока нет
- Hea 1 PERSONAL AND COMMUNITY HEALTHДокумент4 страницыHea 1 PERSONAL AND COMMUNITY HEALTHvaneknekОценок пока нет
- Hope - 1 Grade 11: Exercise For FitnessДокумент11 страницHope - 1 Grade 11: Exercise For FitnessMark Johnson San JuanОценок пока нет